|
Size: 16844
Comment:
|
Size: 2104
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| Post code that demonstrates the use of the interact command in Sage here. It should be easy to just scroll through and paste examples out of here into their own sage notebooks.If you have suggestions on how to improve interact, add them [:interactSuggestions: here] or email [email protected]. | '''WARNING: These are all broken as of Sept 2016. Fixing this is hopefully a high priority. -- William Stein''' |
| Line 5: | Line 5: |
| * [:interact/graph_theory:Graph Theory] * [:interact/calculus:Calculus] * [:interact/diffeq:Differential Equations] * [:interact/linear_algebra:Linear Algebra] * [:interact/algebra:Algebra] * [:interact/number_theory:Number Theory] |
This is a collection of pages demonstrating the use of [[http://sagemath.org/doc/reference/sagenb/notebook/interact.html#sagenb.notebook.interact.interact|the interact command]] in Sage. It should be easy to just scroll through and copy/paste examples into Sage notebooks. If you have suggestions on how to improve interact, add them [[interactSuggestions|here]] or email [email protected] . Of course, your own examples are also welcome! |
| Line 12: | Line 7: |
| == Web applications == | * [[interact/algebra|Algebra]] * [[interact/bio|Bioinformatics]] * [[interact/calculus|Calculus]] * [[interact/diffeq|Differential Equations]] * [[interact/graphics|Drawing Graphics]] * [[interact/dynsys|Dynamical Systems]] * [[interact/fractal|Fractals]] * [[interact/games|Games and Diversions]] * [[interact/geometry|Geometry]] * [[interact/graph_theory|Graph Theory]] * [[interact/linear_algebra|Linear Algebra]] * [[interact/Loop Quantum Gravity|Loop Quantum Gravity]] * [[interact/misc|Miscellaneous]] * [[interact/number_theory|Number Theory]] * [[interact/stats|Statistics/Probability]] * [[interact/topology|Topology]] * [[interact/web|Web Applications]] |
| Line 14: | Line 25: |
| === Stock Market data, fetched from Yahoo and Google === by William Stein |
== Explanatory example: Taylor Series == |
| Line 17: | Line 27: |
| {{{ import urllib |
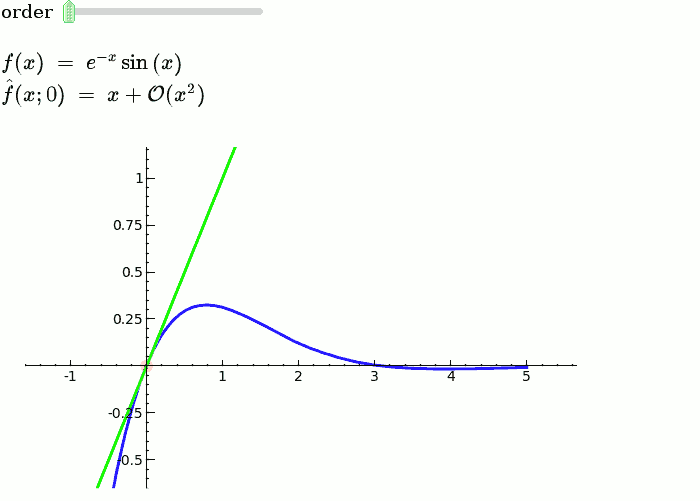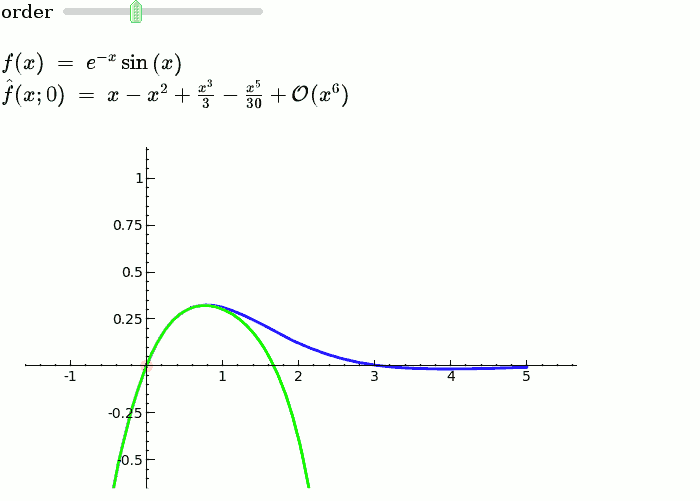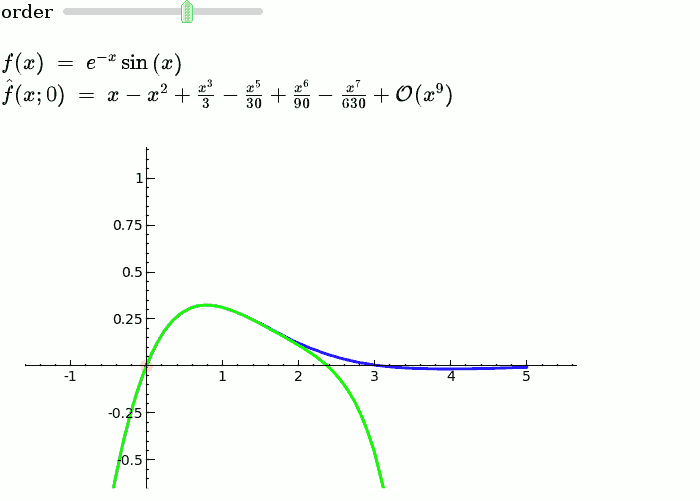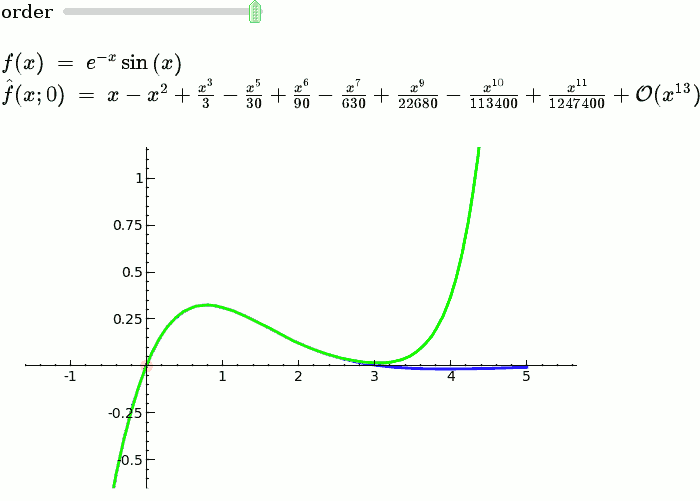
This is the code and a mockup animation of the interact command. It defines a slider, seen on top, that can be dragged. Once dragged, it changes the value of the variable "order" and the whole block of code gets evaluated. This principle can be seen in various examples presented on the pages above! |
| Line 20: | Line 29: |
| class Day: def __init__(self, date, open, high, low, close, volume): self.date = date self.open=float(open); self.high=float(high); self.low=float(low); self.close=float(close) self.volume=int(volume) def __repr__(self): return '%10s %4.2f %4.2f %4.2f %4.2f %10d'%(self.date, self.open, self.high, self.low, self.close, self.volume) class Stock: def __init__(self, symbol): self.symbol = symbol.upper() def __repr__(self): return "%s (%s)"%(self.symbol, self.yahoo()['price']) def yahoo(self): url = 'http://finance.yahoo.com/d/quotes.csv?s=%s&f=%s' % (self.symbol, 'l1c1va2xj1b4j4dyekjm3m4rr5p5p6s7') values = urllib.urlopen(url).read().strip().strip('"').split(',') data = {} data['price'] = values[0] data['change'] = values[1] data['volume'] = values[2] data['avg_daily_volume'] = values[3] data['stock_exchange'] = values[4] data['market_cap'] = values[5] data['book_value'] = values[6] data['ebitda'] = values[7] data['dividend_per_share'] = values[8] data['dividend_yield'] = values[9] data['earnings_per_share'] = values[10] data['52_week_high'] = values[11] data['52_week_low'] = values[12] data['50day_moving_avg'] = values[13] data['200day_moving_avg'] = values[14] data['price_earnings_ratio'] = values[15] data['price_earnings_growth_ratio'] = values[16] data['price_sales_ratio'] = values[17] data['price_book_ratio'] = values[18] data['short_ratio'] = values[19] return data def historical(self): try: return self.__historical except AttributeError: pass symbol = self.symbol def get_data(exchange): name = get_remote_file('http://finance.google.com/finance/historical?q=%s:%s&output=csv'%(exchange, symbol.upper()), verbose=False) return open(name).read() R = get_data('NASDAQ') if "Bad Request" in R: R = get_data("NYSE") R = R.splitlines() headings = R[0].split(',') self.__historical = [] try: for x in reversed(R[1:]): date, opn, high, low, close, volume = x.split(',') self.__historical.append(Day(date, opn,high,low,close,volume)) except ValueError: pass self.__historical = Sequence(self.__historical,cr=True,universe=lambda x:x) return self.__historical def plot_average(self, spline_samples=10): d = self.historical() if len(d) == 0: return text('no historical data at Google Finance about %s'%self.symbol, (0,3)) avg = list(enumerate([(z.high+z.low)/2 for z in d])) P = line(avg) + points(avg, rgbcolor='black', pointsize=4) + \ text(self.symbol, (len(d)*1.05, d[-1].low), horizontal_alignment='right', rgbcolor='black') if spline_samples > 0: k = 250//spline_samples spl = spline([avg[i*k] for i in range(len(d)//k)] + [avg[-1]]) P += plot(spl, (0,len(d)+30), color=(0.7,0.7,0.7)) P.xmax(260) return P def plot_diff(self): d = self.historical() if len(d) == 0: return text('no historical data at Google Finance about %s'%self.symbol, (0,3)) diff = [] for i in range(1, len(d)): z1 = d[i]; z0 = d[i-1] diff.append((i, (z1.high+z1.low)/2 - (z0.high + z0.low)/2)) P = line(diff,thickness=0.5) + points(diff, rgbcolor='black', pointsize=4) + \ text(self.symbol, (len(d)*1.05, 0), horizontal_alignment='right', rgbcolor='black') P.xmax(260) return P symbols = ['bsc', 'vmw', 'sbux', 'aapl', 'amzn', 'goog', 'wfmi', 'msft', 'yhoo', 'ebay', 'java', 'rht', ]; symbols.sort() stocks = dict([(s,Stock(s)) for s in symbols]) |
{{{#!sagecell var('x') x0 = 0 f = sin(x)*e^(-x) p = plot(f,-1,5, thickness=2) dot = point((x0,f(x=x0)),pointsize=80,rgbcolor=(1,0,0)) |
| Line 118: | Line 36: |
| def data(symbol = symbols, other_symbol='', spline_samples=(8,[0..15])): if other_symbol != '': symbol = other_symbol S = Stock(symbol) html('<h1 align=center><font color="darkred">%s</font></h1>'%S) S.plot_average(spline_samples).save('avg.png', figsize=[10,2]) S.plot_diff().save('diff.png', figsize=[10,2]) Y = S.yahoo() k = Y.keys(); k.sort() html('Price during last 52 weeks:<br>Grey line is a spline through %s points (do not take seriously!):<br> <img src="cell://avg.png">'%spline_samples) html('Difference from previous day:<br> <img src="cell://diff.png">') html('<table align=center>' + '\n'.join('<tr><td>%s</td><td>%s</td></tr>'%(k[i], Y[k[i]]) for i in range(len(k))) + '</table>') |
def _(order=(1..12)): ft = f.taylor(x,x0,order) pt = plot(ft,-1, 5, color='green', thickness=2) html('$f(x)\;=\;%s$'%latex(f)) html('$\hat{f}(x;%s)\;=\;%s+\mathcal{O}(x^{%s})$'%(x0,latex(ft),order+1)) show(dot + p + pt, ymin = -.5, ymax = 1) |
| Line 133: | Line 43: |
attachment:stocks.png === CO2 data plot, fetched from NOAA === by Marshall Hampton While support for R is rapidly improving, scipy.stats has a lot of useful stuff too. This only scratches the surface. {{{ import urllib2 as U import scipy.stats as Stat co2data = U.urlopen('ftp://ftp.cmdl.noaa.gov/ccg/co2/trends/co2_mm_mlo.txt').readlines() datalines = [] for a_line in co2data: if a_line.find('Creation:') != -1: cdate = a_line if a_line[0] != '#': temp = a_line.replace('\n','').split(' ') temp = [float(q) for q in temp if q != ''] datalines.append(temp) trdf = RealField(16) @interact def mauna_loa_co2(start_date = slider(1958,2010,1,1958), end_date = slider(1958, 2010,1,2009)): htmls1 = '<h3>CO2 monthly averages at Mauna Loa (interpolated), from NOAA/ESRL data</h3>' htmls2 = '<h4>'+cdate+'</h4>' sel_data = [[q[2],q[4]] for q in datalines if start_date < q[2] < end_date] c_max = max([q[1] for q in sel_data]) c_min = min([q[1] for q in sel_data]) slope, intercept, r, ttprob, stderr = Stat.linregress(sel_data) html(htmls1+htmls2+'<h4>Linear regression slope: ' + str(trdf(slope)) + ' ppm/year; correlation coefficient: ' + str(trdf(r)) + '</h4>') var('x,y') show(list_plot(sel_data, plotjoined=True, rgbcolor=(1,0,0)) + plot(slope*x+intercept,start_date,end_date), xmin = start_date, ymin = c_min-2, axes = True, xmax = end_date, ymax = c_max+3, frame = False) }}} attachment:co2c.png === Pie Chart from the Google Chart API === by Harald Schilly {{{ # Google Chart API: http://code.google.com/apis/chart import urllib2 as inet from pylab import imshow @interact def gChart(title="Google Chart API plots Pie Charts!", color1=Color('purple'), color2=Color('black'), color3=Color('yellow'), val1=slider(0,1,.05,.5), val2=slider(0,1,.05,.3), val3=slider(0,1,.05,0.1), label=("Maths Physics Chemistry")): url = "http://chart.apis.google.com/chart?cht=p3&chs=600x300" url += '&chtt=%s&chts=000000,25'%title.replace(" ","+") url += '&chco=%s'%(','.join([color1.html_color()[1:],color2.html_color()[1:],color3.html_color()[1:]])) url += '&chl=%s'%label.replace(" ","|") url += '&chd=t:%s'%(','.join(map(str,[val1,val2,val3]))) print url html('<div style="border:3px dashed;text-align:center;padding:50px 0 50px 0"><img src="%s"></div>'%url) }}} attachment:interact_with_google_chart_api.png == Bioinformatics == === Web app: protein browser === by Marshall Hampton (tested by William Stein) {{{ import urllib2 as U @interact def protein_browser(GenBank_ID = input_box('165940577', type = str), file_type = selector([(1,'fasta'),(2,'GenPept')])): if file_type == 2: gen_str = 'http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&sendto=t&id=' else: gen_str = 'http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&sendto=t&dopt=fasta&id=' f = U.urlopen(gen_str + GenBank_ID) g = f.read() f.close() html(g) }}} attachment:biobrowse.png === Coalescent simulator === by Marshall Hampton {{{ def next_gen(x, selection=1.0): '''Creates the next generation from the previous; also returns parent-child indexing list''' next_x = [] for ind in range(len(x)): if random() < (1 + selection)/len(x): rind = 0 else: rind = int(round(random()*(len(x)-1)+1/2)) next_x.append((x[rind],rind)) next_x.sort() return [[x[0] for x in next_x],[x[1] for x in next_x]] def coal_plot(some_data): '''Creates a graphics object from coalescent data''' gens = some_data[0] inds = some_data[1] gen_lines = line([[0,0]]) pts = Graphics() ngens = len(gens) gen_size = len(gens[0]) for x in range(gen_size): pts += point((x,ngens-1), hue = gens[0][x]/float(gen_size*1.1)) p_frame = line([[-.5,-.5],[-.5,ngens-.5], [gen_size-.5,ngens-.5], [gen_size-.5,-.5], [-.5,-.5]]) for g in range(1,ngens): for x in range(gen_size): old_x = inds[g-1][x] gen_lines += line([[x,ngens-g-1],[old_x,ngens-g]], hue = gens[g-1][old_x]/float(gen_size*1.1)) pts += point((x,ngens-g-1), hue = gens[g][x]/float(gen_size*1.1)) return pts+gen_lines+p_frame d_field = RealField(10) @interact def coalescents(pop_size = slider(2,100,1,15,'Population size'), selection = slider(-1,1,.1,0, 'Selection for first taxon'), s = selector(['Again!'], label='Refresh', buttons=True)): print 'Population size: ' + str(pop_size) print 'Selection coefficient for first taxon: ' + str(d_field(selection)) start = [i for i in range(pop_size)] gens = [start] inds = [] while gens[-1][0] != gens[-1][-1]: g_index = len(gens) - 1 n_gen = next_gen(gens[g_index], selection = selection) gens.append(n_gen[0]) inds.append(n_gen[1]) coal_data1 = [gens,inds] print 'Generations until coalescence: ' + str(len(gens)) show(coal_plot(coal_data1), axes = False, figsize = [8,4.0*len(gens)/pop_size], ymax = len(gens)-1) }}} attachment:coalescent.png == Miscellaneous Graphics == === Catalog of 3D Parametric Plots === {{{ var('u,v') plots = ['Two Interlinked Tori', 'Star of David', 'Double Heart', 'Heart', 'Green bowtie', "Boy's Surface", "Maeder's Owl", 'Cross cap'] plots.sort() @interact def _(example=selector(plots, buttons=True, nrows=2), tachyon=("Raytrace", False), frame = ('Frame', False), opacity=(1,(0.1,1))): url = '' if example == 'Two Interlinked Tori': f1 = (4+(3+cos(v))*sin(u), 4+(3+cos(v))*cos(u), 4+sin(v)) f2 = (8+(3+cos(v))*cos(u), 3+sin(v), 4+(3+cos(v))*sin(u)) p1 = parametric_plot3d(f1, (u,0,2*pi), (v,0,2*pi), color="red", opacity=opacity) p2 = parametric_plot3d(f2, (u,0,2*pi), (v,0,2*pi), color="blue",opacity=opacity) P = p1 + p2 elif example == 'Star of David': f_x = cos(u)*cos(v)*(abs(cos(3*v/4))^500 + abs(sin(3*v/4))^500)^(-1/260)*(abs(cos(4*u/4))^200 + abs(sin(4*u/4))^200)^(-1/200) f_y = cos(u)*sin(v)*(abs(cos(3*v/4))^500 + abs(sin(3*v/4))^500)^(-1/260)*(abs(cos(4*u/4))^200 + abs(sin(4*u/4))^200)^(-1/200) f_z = sin(u)*(abs(cos(4*u/4))^200 + abs(sin(4*u/4))^200)^(-1/200) P = parametric_plot3d([f_x, f_y, f_z], (u, -pi, pi), (v, 0, 2*pi),opacity=opacity) elif example == 'Double Heart': f_x = ( abs(v) - abs(u) - abs(tanh((1/sqrt(2))*u)/(1/sqrt(2))) + abs(tanh((1/sqrt(2))*v)/(1/sqrt(2))) )*sin(v) f_y = ( abs(v) - abs(u) - abs(tanh((1/sqrt(2))*u)/(1/sqrt(2))) - abs(tanh((1/sqrt(2))*v)/(1/sqrt(2))) )*cos(v) f_z = sin(u)*(abs(cos(4*u/4))^1 + abs(sin(4*u/4))^1)^(-1/1) P = parametric_plot3d([f_x, f_y, f_z], (u, 0, pi), (v, -pi, pi),opacity=opacity) elif example == 'Heart': f_x = cos(u)*(4*sqrt(1-v^2)*sin(abs(u))^abs(u)) f_y = sin(u) *(4*sqrt(1-v^2)*sin(abs(u))^abs(u)) f_z = v P = parametric_plot3d([f_x, f_y, f_z], (u, -pi, pi), (v, -1, 1), frame=False, color="red",opacity=opacity) elif example == 'Green bowtie': f_x = sin(u) / (sqrt(2) + sin(v)) f_y = sin(u) / (sqrt(2) + cos(v)) f_z = cos(u) / (1 + sqrt(2)) P = parametric_plot3d([f_x, f_y, f_z], (u, -pi, pi), (v, -pi, pi), frame=False, color="green",opacity=opacity) elif example == "Boy's Surface": url = "http://en.wikipedia.org/wiki/Boy's_surface" fx = 2/3* (cos(u)* cos(2*v) + sqrt(2)* sin(u)* cos(v))* cos(u) / (sqrt(2) - sin(2*u)* sin(3*v)) fy = 2/3* (cos(u)* sin(2*v) - sqrt(2)* sin(u)* sin(v))* cos(u) / (sqrt(2) - sin(2*u)* sin(3*v)) fz = sqrt(2)* cos(u)* cos(u) / (sqrt(2) - sin(2*u)* sin(3*v)) P = parametric_plot3d([fx, fy, fz], (u, -2*pi, 2*pi), (v, 0, pi), plot_points = [90,90], frame=False, color="orange",opacity=opacity) elif example == "Maeder's Owl": fx = v *cos(u) - 0.5* v^2 * cos(2* u) fy = -v *sin(u) - 0.5* v^2 * sin(2* u) fz = 4 *v^1.5 * cos(3 *u / 2) / 3 P = parametric_plot3d([fx, fy, fz], (u, -2*pi, 2*pi), (v, 0, 1),plot_points = [90,90], frame=False, color="purple",opacity=opacity) elif example =='Cross cap': url = 'http://en.wikipedia.org/wiki/Cross-cap' fx = (1+cos(v))*cos(u) fy = (1+cos(v))*sin(u) fz = -tanh((2/3)*(u-pi))*sin(v) P = parametric_plot3d([fx, fy, fz], (u, 0, 2*pi), (v, 0, 2*pi), frame=False, color="red",opacity=opacity) else: print "Bug selecting plot?" return html('<h2>%s</h2>'%example) if url: html('<h3><a target="_new" href="%s">%s</a></h3>'%(url,url)) show(P, viewer='tachyon' if tachyon else 'jmol', frame=frame) }}} attachment:parametricplot3d.png === Interactive rotatable raytracing with Tachyon3d === {{{ C = cube(color=['red', 'green', 'blue'], aspect_ratio=[1,1,1], viewer='tachyon') + sphere((1,0,0),0.2) @interact def example(theta=(0,2*pi), phi=(0,2*pi), zoom=(1,(1,4))): show(C.rotate((0,0,1), theta).rotate((0,1,0),phi), zoom=zoom) }}} attachment:tachyonrotate.png === Interactive 3d plotting === {{{ var('x,y') @interact def example(clr=Color('orange'), f=4*x*exp(-x^2-y^2), xrange='(-2, 2)', yrange='(-2,2)', zrot=(0,pi), xrot=(0,pi), zoom=(1,(1/2,3)), square_aspect=('Square Frame', False), tachyon=('Ray Tracer', True)): xmin, xmax = sage_eval(xrange); ymin, ymax = sage_eval(yrange) P = plot3d(f, (x, xmin, xmax), (y, ymin, ymax), color=clr) html('<h1>Plot of $f(x,y) = %s$</h1>'%latex(f)) aspect_ratio = [1,1,1] if square_aspect else [1,1,1/2] show(P.rotate((0,0,1), -zrot).rotate((1,0,0),xrot), viewer='tachyon' if tachyon else 'jmol', figsize=6, zoom=zoom, frame=False, frame_aspect_ratio=aspect_ratio) }}} attachment:tachyonplot3d.png [[Anchor(eggpaint)]] === Somewhat Silly Egg Painter === by Marshall Hampton (refereed by William Stein) {{{ var('s,t') g(s) = ((0.57496*sqrt(121 - 16.0*s^2))/sqrt(10.+ s)) def P(color, rng): return parametric_plot3d((cos(t)*g(s), sin(t)*g(s), s), (s,rng[0],rng[1]), (t,0,2*pi), plot_points = [150,150], rgbcolor=color, frame = False, opacity = 1) colorlist = ['red','blue','red','blue'] @interact def _(band_number = selector(range(1,5)), current_color = Color('red')): html('<h1 align=center>Egg Painter</h1>') colorlist[band_number-1] = current_color egg = sum([P(colorlist[i],[-2.75+5.5*(i/4),-2.75+5.5*(i+1)/4]) for i in range(4)]) show(egg) }}} attachment:eggpaint.png ===Miscellaneous=== Profile a snippet of code {{{ html('<h2>Profile the given input</h2>') import cProfile; import profile @interact def _(cmd = ("Statement", '2 + 2'), do_preparse=("Preparse?", True), cprof =("cProfile?", False)): if do_preparse: cmd = preparse(cmd) print "<html>" # trick to avoid word wrap if cprof: cProfile.run(cmd) else: profile.run(cmd) print "</html>" }}} attachment:profile.png |
{{attachment:taylor_series_animated.gif}} |
Sage Interactions
WARNING: These are all broken as of Sept 2016. Fixing this is hopefully a high priority. -- William Stein
This is a collection of pages demonstrating the use of the interact command in Sage. It should be easy to just scroll through and copy/paste examples into Sage notebooks. If you have suggestions on how to improve interact, add them here or email [email protected] . Of course, your own examples are also welcome!
Explanatory example: Taylor Series
This is the code and a mockup animation of the interact command. It defines a slider, seen on top, that can be dragged. Once dragged, it changes the value of the variable "order" and the whole block of code gets evaluated. This principle can be seen in various examples presented on the pages above!