|
Size: 11124
Comment:
|
Size: 2619
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| Post code that demonstrates the use of the interact command in Sage here. It should be easy to just scroll through and paste examples out of here into their own sage notebooks.If you have suggestions on how to improve interact, add them [:interactSuggestions: here] or email sage-support@googlegroups.com. | This is a collection of pages demonstrating the use of the **interact** command in Sage. It should be easy to just scroll through and copy/paste examples into Sage notebooks. If you have suggestions on how to improve interact, add them [[interact/Suggestions|here]] or email the sage-support mailing list. Of course, your own examples are also welcome! |
| Line 5: | Line 8: |
| * [:interact/graph_theory:Graph Theory] * [:interact/calculus:Calculus] * [:interact/diffeq:Differential Equations] * [:interact/linear_algebra:Linear Algebra] * [:interact/algebra:Algebra] * [:interact/number_theory:Number Theory] * [:interact/web:Web Applications] |
Documentation links: |
| Line 13: | Line 10: |
| == Bioinformatics == | * [[http://doc.sagemath.org/html/en/reference/repl/sage/repl/ipython_kernel/interact.html| interacts in the Jupyter notebook]] (see this page and the two following ones) * [[https://github.com/sagemath/sagenb/blob/master/sagenb/notebook/interact.py|interacts in the legacy SageNB notebook]] (many helpful examples) * [[https://github.com/sagemath/sagecell/blob/master/interact_compatibility.py|Sage Cell Server implementation]] * [[https://github.com/sagemathinc/cocalc/blob/master/src/smc_sagews/smc_sagews/sage_salvus.py#L348|CoCalc Sage worksheet implementation]] |
| Line 15: | Line 15: |
| === Web app: protein browser === by Marshall Hampton (tested by William Stein) {{{ import urllib2 as U @interact def protein_browser(GenBank_ID = input_box('165940577', type = str), file_type = selector([(1,'fasta'),(2,'GenPept')])): if file_type == 2: gen_str = 'http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&sendto=t&id=' else: gen_str = 'http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&sendto=t&dopt=fasta&id=' f = U.urlopen(gen_str + GenBank_ID) g = f.read() f.close() html(g) }}} attachment:biobrowse.png |
Examples: |
| Line 32: | Line 17: |
| === Coalescent simulator === by Marshall Hampton {{{ def next_gen(x, selection=1.0): '''Creates the next generation from the previous; also returns parent-child indexing list''' next_x = [] for ind in range(len(x)): if random() < (1 + selection)/len(x): rind = 0 else: rind = int(round(random()*(len(x)-1)+1/2)) next_x.append((x[rind],rind)) next_x.sort() return [[x[0] for x in next_x],[x[1] for x in next_x]] def coal_plot(some_data): '''Creates a graphics object from coalescent data''' gens = some_data[0] inds = some_data[1] gen_lines = line([[0,0]]) pts = Graphics() ngens = len(gens) gen_size = len(gens[0]) for x in range(gen_size): pts += point((x,ngens-1), hue = gens[0][x]/float(gen_size*1.1)) p_frame = line([[-.5,-.5],[-.5,ngens-.5], [gen_size-.5,ngens-.5], [gen_size-.5,-.5], [-.5,-.5]]) for g in range(1,ngens): for x in range(gen_size): old_x = inds[g-1][x] gen_lines += line([[x,ngens-g-1],[old_x,ngens-g]], hue = gens[g-1][old_x]/float(gen_size*1.1)) pts += point((x,ngens-g-1), hue = gens[g][x]/float(gen_size*1.1)) return pts+gen_lines+p_frame d_field = RealField(10) @interact def coalescents(pop_size = slider(2,100,1,15,'Population size'), selection = slider(-1,1,.1,0, 'Selection for first taxon'), s = selector(['Again!'], label='Refresh', buttons=True)): print 'Population size: ' + str(pop_size) print 'Selection coefficient for first taxon: ' + str(d_field(selection)) start = [i for i in range(pop_size)] gens = [start] inds = [] while gens[-1][0] != gens[-1][-1]: g_index = len(gens) - 1 n_gen = next_gen(gens[g_index], selection = selection) gens.append(n_gen[0]) inds.append(n_gen[1]) coal_data1 = [gens,inds] print 'Generations until coalescence: ' + str(len(gens)) show(coal_plot(coal_data1), axes = False, figsize = [8,4.0*len(gens)/pop_size], ymax = len(gens)-1) }}} attachment:coalescent.png |
* [[interact/algebra|Algebra]] * [[interact/bio|Bioinformatics]] * [[interact/calculus|Calculus]] * [[interact/cryptography|Cryptography]] * [[interact/diffeq|Differential Equations]] * [[interact/graphics|Drawing Graphics]] * [[interact/dynsys|Dynamical Systems]] * [[interact/fractal|Fractals]] * [[interact/games|Games and Diversions]] * [[interact/geometry|Geometry]] * [[interact/graph_theory|Graph Theory]] * [[interact/linear_algebra|Linear Algebra]] * [[interact/Loop Quantum Gravity|Loop Quantum Gravity]] * [[interact/misc|Miscellaneous]] * [[interact/number_theory|Number Theory]] * [[interact/stats|Statistics/Probability]] * [[interact/topology|Topology]] * [[interact/web|Web Applications]] |
| Line 82: | Line 36: |
| == Miscellaneous Graphics == | == Explanatory example: Taylor Series == |
| Line 84: | Line 38: |
| === Catalog of 3D Parametric Plots === {{{ var('u,v') plots = ['Two Interlinked Tori', 'Star of David', 'Double Heart', 'Heart', 'Green bowtie', "Boy's Surface", "Maeder's Owl", 'Cross cap'] plots.sort() |
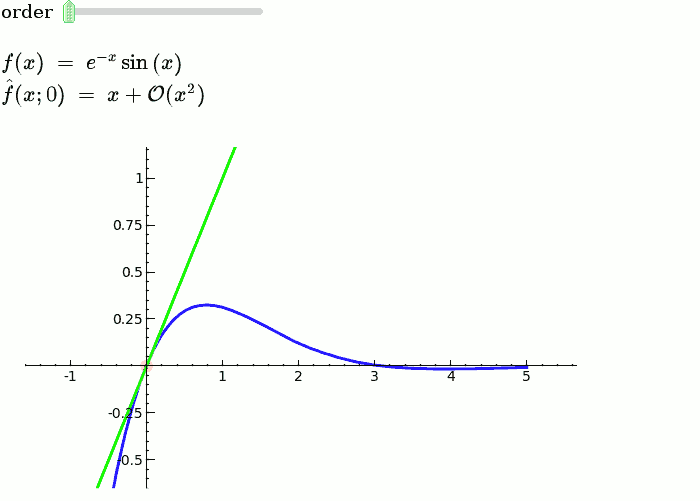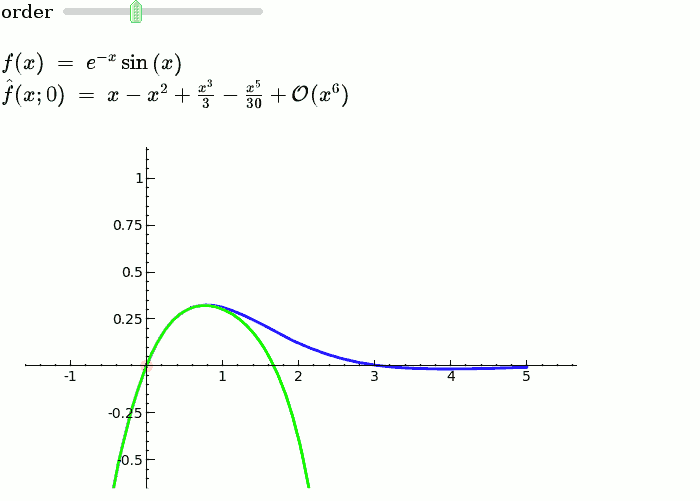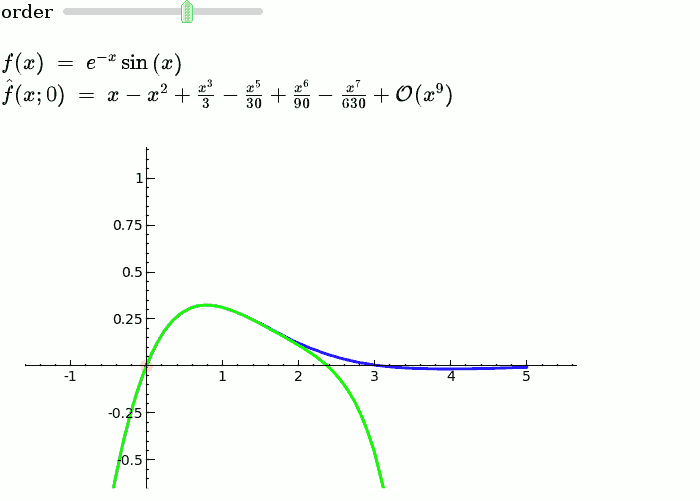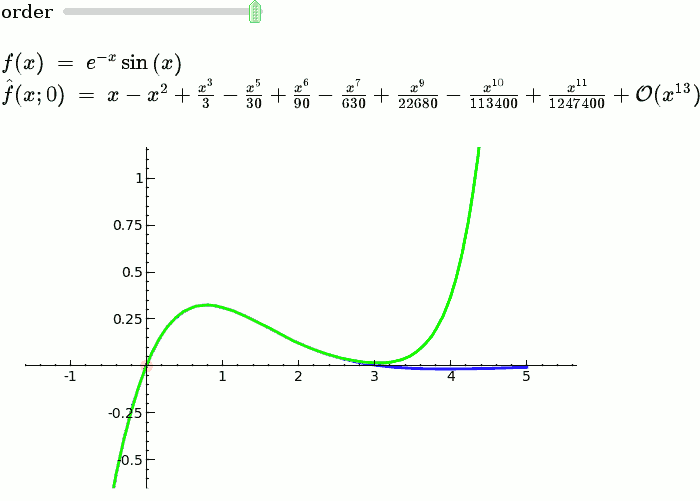
This is the code and a mockup animation of the interact command. It defines a slider, seen on top, that can be dragged. Once dragged, it changes the value of the variable "order" and the whole block of code gets evaluated. This principle can be seen in various examples presented on the pages above! {{{#!sagecell x = SR.var('x') x0 = 0 f = sin(x) * e^(-x) p = plot(f, -1, 5, thickness=2) dot = point((x0, f(x=x0)), pointsize=80, rgbcolor=(1, 0, 0)) |
| Line 93: | Line 48: |
| def _(example=selector(plots, buttons=True, nrows=2), tachyon=("Raytrace", False), frame = ('Frame', False), opacity=(1,(0.1,1))): url = '' if example == 'Two Interlinked Tori': f1 = (4+(3+cos(v))*sin(u), 4+(3+cos(v))*cos(u), 4+sin(v)) f2 = (8+(3+cos(v))*cos(u), 3+sin(v), 4+(3+cos(v))*sin(u)) p1 = parametric_plot3d(f1, (u,0,2*pi), (v,0,2*pi), color="red", opacity=opacity) p2 = parametric_plot3d(f2, (u,0,2*pi), (v,0,2*pi), color="blue",opacity=opacity) P = p1 + p2 elif example == 'Star of David': f_x = cos(u)*cos(v)*(abs(cos(3*v/4))^500 + abs(sin(3*v/4))^500)^(-1/260)*(abs(cos(4*u/4))^200 + abs(sin(4*u/4))^200)^(-1/200) f_y = cos(u)*sin(v)*(abs(cos(3*v/4))^500 + abs(sin(3*v/4))^500)^(-1/260)*(abs(cos(4*u/4))^200 + abs(sin(4*u/4))^200)^(-1/200) f_z = sin(u)*(abs(cos(4*u/4))^200 + abs(sin(4*u/4))^200)^(-1/200) P = parametric_plot3d([f_x, f_y, f_z], (u, -pi, pi), (v, 0, 2*pi),opacity=opacity) elif example == 'Double Heart': f_x = ( abs(v) - abs(u) - abs(tanh((1/sqrt(2))*u)/(1/sqrt(2))) + abs(tanh((1/sqrt(2))*v)/(1/sqrt(2))) )*sin(v) f_y = ( abs(v) - abs(u) - abs(tanh((1/sqrt(2))*u)/(1/sqrt(2))) - abs(tanh((1/sqrt(2))*v)/(1/sqrt(2))) )*cos(v) f_z = sin(u)*(abs(cos(4*u/4))^1 + abs(sin(4*u/4))^1)^(-1/1) P = parametric_plot3d([f_x, f_y, f_z], (u, 0, pi), (v, -pi, pi),opacity=opacity) elif example == 'Heart': f_x = cos(u)*(4*sqrt(1-v^2)*sin(abs(u))^abs(u)) f_y = sin(u) *(4*sqrt(1-v^2)*sin(abs(u))^abs(u)) f_z = v P = parametric_plot3d([f_x, f_y, f_z], (u, -pi, pi), (v, -1, 1), frame=False, color="red",opacity=opacity) elif example == 'Green bowtie': f_x = sin(u) / (sqrt(2) + sin(v)) f_y = sin(u) / (sqrt(2) + cos(v)) f_z = cos(u) / (1 + sqrt(2)) P = parametric_plot3d([f_x, f_y, f_z], (u, -pi, pi), (v, -pi, pi), frame=False, color="green",opacity=opacity) elif example == "Boy's Surface": url = "http://en.wikipedia.org/wiki/Boy's_surface" fx = 2/3* (cos(u)* cos(2*v) + sqrt(2)* sin(u)* cos(v))* cos(u) / (sqrt(2) - sin(2*u)* sin(3*v)) fy = 2/3* (cos(u)* sin(2*v) - sqrt(2)* sin(u)* sin(v))* cos(u) / (sqrt(2) - sin(2*u)* sin(3*v)) fz = sqrt(2)* cos(u)* cos(u) / (sqrt(2) - sin(2*u)* sin(3*v)) P = parametric_plot3d([fx, fy, fz], (u, -2*pi, 2*pi), (v, 0, pi), plot_points = [90,90], frame=False, color="orange",opacity=opacity) elif example == "Maeder's Owl": fx = v *cos(u) - 0.5* v^2 * cos(2* u) fy = -v *sin(u) - 0.5* v^2 * sin(2* u) fz = 4 *v^1.5 * cos(3 *u / 2) / 3 P = parametric_plot3d([fx, fy, fz], (u, -2*pi, 2*pi), (v, 0, 1),plot_points = [90,90], frame=False, color="purple",opacity=opacity) elif example =='Cross cap': url = 'http://en.wikipedia.org/wiki/Cross-cap' fx = (1+cos(v))*cos(u) fy = (1+cos(v))*sin(u) fz = -tanh((2/3)*(u-pi))*sin(v) P = parametric_plot3d([fx, fy, fz], (u, 0, 2*pi), (v, 0, 2*pi), frame=False, color="red",opacity=opacity) else: print "Bug selecting plot?" return html('<h2>%s</h2>'%example) if url: html('<h3><a target="_new" href="%s">%s</a></h3>'%(url,url)) show(P, viewer='tachyon' if tachyon else 'jmol', frame=frame) |
def _(order=slider([1 .. 12])): ft = f.taylor(x, x0, order) pt = plot(ft, -1, 5, color='green', thickness=2) pretty_print(html(r'$f(x)\;=\;%s$' % latex(f))) pretty_print(html(r'$\hat{f}(x;%s)\;=\;%s+\mathcal{O}(x^{%s})$' % (x0, latex(ft), order+1))) show(dot + p + pt, ymin=-.5, ymax=1) |
| Line 150: | Line 55: |
attachment:parametricplot3d.png === Interactive rotatable raytracing with Tachyon3d === {{{ C = cube(color=['red', 'green', 'blue'], aspect_ratio=[1,1,1], viewer='tachyon') + sphere((1,0,0),0.2) @interact def example(theta=(0,2*pi), phi=(0,2*pi), zoom=(1,(1,4))): show(C.rotate((0,0,1), theta).rotate((0,1,0),phi), zoom=zoom) }}} attachment:tachyonrotate.png === Interactive 3d plotting === {{{ var('x,y') @interact def example(clr=Color('orange'), f=4*x*exp(-x^2-y^2), xrange='(-2, 2)', yrange='(-2,2)', zrot=(0,pi), xrot=(0,pi), zoom=(1,(1/2,3)), square_aspect=('Square Frame', False), tachyon=('Ray Tracer', True)): xmin, xmax = sage_eval(xrange); ymin, ymax = sage_eval(yrange) P = plot3d(f, (x, xmin, xmax), (y, ymin, ymax), color=clr) html('<h1>Plot of $f(x,y) = %s$</h1>'%latex(f)) aspect_ratio = [1,1,1] if square_aspect else [1,1,1/2] show(P.rotate((0,0,1), -zrot).rotate((1,0,0),xrot), viewer='tachyon' if tachyon else 'jmol', figsize=6, zoom=zoom, frame=False, frame_aspect_ratio=aspect_ratio) }}} attachment:tachyonplot3d.png [[Anchor(eggpaint)]] === Somewhat Silly Egg Painter === by Marshall Hampton (refereed by William Stein) {{{ var('s,t') g(s) = ((0.57496*sqrt(121 - 16.0*s^2))/sqrt(10.+ s)) def P(color, rng): return parametric_plot3d((cos(t)*g(s), sin(t)*g(s), s), (s,rng[0],rng[1]), (t,0,2*pi), plot_points = [150,150], rgbcolor=color, frame = False, opacity = 1) colorlist = ['red','blue','red','blue'] @interact def _(band_number = selector(range(1,5)), current_color = Color('red')): html('<h1 align=center>Egg Painter</h1>') colorlist[band_number-1] = current_color egg = sum([P(colorlist[i],[-2.75+5.5*(i/4),-2.75+5.5*(i+1)/4]) for i in range(4)]) show(egg) }}} attachment:eggpaint.png == Miscellaneous == == Profile a snippet of code == {{{ html('<h2>Profile the given input</h2>') import cProfile; import profile @interact def _(cmd = ("Statement", '2 + 2'), do_preparse=("Preparse?", True), cprof =("cProfile?", False)): if do_preparse: cmd = preparse(cmd) print "<html>" # trick to avoid word wrap if cprof: cProfile.run(cmd) else: profile.run(cmd) print "</html>" }}} attachment:profile.png === Evaluate a bit of code in a given system === by William Stein (there is no way yet to make the text box big): {{{ @interact def _(system=selector([('sage0', 'Sage'), ('gp', 'PARI'), ('magma', 'Magma')]), code='2+2'): print globals()[system].eval(code) }}} attachment:evalsys.png === A Random Walk === by William Stein {{{ html('<h1>A Random Walk</h1>') vv = []; nn = 0 @interact def foo(pts = checkbox(True, "Show points"), refresh = checkbox(False, "New random walk every time"), steps = (50,(10..500))): # We cache the walk in the global variable vv, so that # checking or unchecking the points checkbox doesn't change # the random walk. html("<h2>%s steps</h2>"%steps) global vv if refresh or len(vv) == 0: s = 0; v = [(0,0)] for i in range(steps): s += random() - 0.5 v.append((i, s)) vv = v elif len(vv) != steps: # Add or subtract some points s = vv[-1][1]; j = len(vv) for i in range(steps - len(vv)): s += random() - 0.5 vv.append((i+j,s)) v = vv[:steps] else: v = vv L = line(v, rgbcolor='#4a8de2') if pts: L += points(v, pointsize=10, rgbcolor='red') show(L, xmin=0, figsize=[8,3]) }}} attachment:randomwalk.png === 3D Random Walk === {{{ @interact def rwalk3d(n=(50,1000), frame=True): pnt = [0,0,0] v = [copy(pnt)] for i in range(n): pnt[0] += random()-0.5 pnt[1] += random()-0.5 pnt[2] += random()-0.5 v.append(copy(pnt)) show(line3d(v,color='black'),aspect_ratio=[1,1,1],frame=frame) }}} attachment:randomwalk3d.png |
{{attachment:taylor_series_animated.gif}} |
Sage Interactions
This is a collection of pages demonstrating the use of the **interact** command in Sage. It should be easy to just scroll through and copy/paste examples into Sage notebooks. If you have suggestions on how to improve interact, add them here or email the sage-support mailing list. Of course, your own examples are also welcome!
Documentation links:
interacts in the Jupyter notebook (see this page and the two following ones)
interacts in the legacy SageNB notebook (many helpful examples)
Examples:
Explanatory example: Taylor Series
This is the code and a mockup animation of the interact command. It defines a slider, seen on top, that can be dragged. Once dragged, it changes the value of the variable "order" and the whole block of code gets evaluated. This principle can be seen in various examples presented on the pages above!