|
Size: 33150
Comment:
|
Size: 1905
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| Post code that demonstrates the use of the interact command in Sage here. It should be easy for people to just scroll through and paste examples out of here into their own sage notebooks. | This is a collection of pages demonstrating the use of [[http://sagemath.org/doc/reference/sagenb/notebook/interact.html#sagenb.notebook.interact.interact|the interact command]] in Sage. It should be easy to just scroll through and copy/paste examples into sage notebooks. If you have suggestions on how to improve interact, add them [[interactSuggestions|here]] or email [email protected] . Of course, your own examples are also welcome! |
| Line 5: | Line 5: |
| We'll likely restructure and reorganize this once we have some nontrivial content and get a sense of how it is laid out. If you have suggestions on how to improve interact, add them [:interactSuggestions: here] or email [email protected]. | * [[interact/graph_theory|Graph Theory]] * [[interact/fractal|Fractals]] * [[interact/calculus|Calculus]] * [[interact/diffeq|Differential Equations]] * [[interact/dynsys|Dynamical Systems]] * [[interact/linear_algebra|Linear Algebra]] * [[interact/algebra|Algebra]] * [[interact/number_theory|Number Theory]] * [[interact/web|Web Applications]] * [[interact/bio|Bioinformatics]] * [[interact/stats|Statistics/Probability]] * [[interact/geometry|Geometry]] * [[interact/graphics|Drawing Graphics]] * [[interact/games|Games and Diversions]] * [[interact/misc|Miscellaneous]] |
| Line 7: | Line 21: |
| [[TableOfContents]] | == Explanatory example: Taylor Series == |
| Line 9: | Line 23: |
| == Miscellaneous == | This is the code and a mockup animation of the interact command. It defines a slider, seen on top, that can be dragged. Once dragged, it changes the value of the variable "order" and the whole block of code gets evaluated. This principle can be seen in various examples presented on the pages above! |
| Line 11: | Line 25: |
| === Evaluate a bit of code in a given system === by William Stein (there is no way yet to make the text box big): {{{ |
{{{#!python numbers=none var('x') x0 = 0 f = sin(x)*e^(-x) p = plot(f,-1,5, thickness=2) dot = point((x0,f(x=x0)),pointsize=80,rgbcolor=(1,0,0)) |
| Line 17: | Line 32: |
| def _(system=selector([('sage0', 'Sage'), ('gp', 'PARI'), ('magma', 'Magma')]), code='2+2'): print globals()[system].eval(code) |
def _(order=(1..12)): ft = f.taylor(x,x0,order) pt = plot(ft,-1, 5, color='green', thickness=2) html('$f(x)\;=\;%s$'%latex(f)) html('$\hat{f}(x;%s)\;=\;%s+\mathcal{O}(x^{%s})$'%(x0,latex(ft),order+1)) show(dot + p + pt, ymin = -.5, ymax = 1) |
| Line 20: | Line 39: |
attachment:evalsys.png == Graph Theory == === Automorphism Groups of some Graphs === by William Stein (I spent less than five minutes on this): {{{ @interact def _(graph=['CycleGraph', 'CubeGraph', 'RandomGNP'], n=selector([1..10],nrows=1), p=selector([10,20,..,100],nrows=1)): print graph if graph == 'CycleGraph': print "n (=%s): number of vertices"%n G = graphs.CycleGraph(n) elif graph == 'CubeGraph': if n > 8: print "n reduced to 8" n = 8 print "n (=%s): dimension"%n G = graphs.CubeGraph(n) elif graph == 'RandomGNP': print "n (=%s) vertices"%n print "p (=%s%%) probability"%p G = graphs.RandomGNP(n, p/100.0) print G.automorphism_group() show(plot(G)) }}} attachment:autograph.png == Calculus == === A contour map and 3d plot of two inverse distance functions === by William Stein {{{ @interact def _(q1=(-1,(-3,3)), q2=(-2,(-3,3)), cmap=['autumn', 'bone', 'cool', 'copper', 'gray', 'hot', 'hsv', 'jet', 'pink', 'prism', 'spring', 'summer', 'winter']): x,y = var('x,y') f = q1/sqrt((x+1)^2 + y^2) + q2/sqrt((x-1)^2+(y+0.5)^2) C = contour_plot(f, (-2,2), (-2,2), plot_points=30, contours=15, cmap=cmap) show(C, figsize=3, aspect_ratio=1) show(plot3d(f, (x,-2,2), (y,-2,2)), figsize=5, viewer='tachyon') }}} attachment:mountains.png === A simple tangent line grapher === by Marshall Hampton {{{ html('<h2>Tangent line grapher</h2>') @interact def tangent_line(f = input_box(default=sin(x)), xbegin = slider(0,10,1/10,0), xend = slider(0,10,1/10,10), x0 = slider(0, 1, 1/100, 1/2)): prange = [xbegin, xend] x0i = xbegin + x0*(xend-xbegin) var('x') df = diff(f) tanf = f(x0i) + df(x0i)*(x-x0i) fplot = plot(f, prange[0], prange[1]) print 'Tangent line is y = ' + tanf._repr_() tanplot = plot(tanf, prange[0], prange[1], rgbcolor = (1,0,0)) fmax = f.find_maximum_on_interval(prange[0], prange[1])[0] fmin = f.find_minimum_on_interval(prange[0], prange[1])[0] show(fplot + tanplot, xmin = prange[0], xmax = prange[1], ymax = fmax, ymin = fmin) }}} attachment:tangents.png === Function tool === Enter symbolic functions $f$, $g$, and $a$, a range, then click the appropriate button to compute and plot some combination of $f$, $g$, and $a$ along with $f$ and $g$. This is inspired by the Matlab funtool GUI. {{{ x = var('x') @interact def _(f=sin(x), g=cos(x), xrange=input_box((0,1)), yrange='auto', a=1, action=selector(['f', 'df/dx', 'int f', 'num f', 'den f', '1/f', 'finv', 'f+a', 'f-a', 'f*a', 'f/a', 'f^a', 'f(x+a)', 'f(x*a)', 'f+g', 'f-g', 'f*g', 'f/g', 'f(g)'], width=15, nrows=5, label="h = "), do_plot = ("Draw Plots", True)): try: f = SR(f); g = SR(g); a = SR(a) except TypeError, msg: print msg[-200:] print "Unable to make sense of f,g, or a as symbolic expressions." return if not (isinstance(xrange, tuple) and len(xrange) == 2): xrange = (0,1) h = 0; lbl = '' if action == 'f': h = f lbl = 'f' elif action == 'df/dx': h = f.derivative(x) lbl = '\\frac{df}{dx}' elif action == 'int f': h = f.integrate(x) lbl = '\\int f dx' elif action == 'num f': h = f.numerator() lbl = '\\text{numer(f)}' elif action == 'den f': h = f.denominator() lbl = '\\text{denom(f)}' elif action == '1/f': h = 1/f lbl = '\\frac{1}{f}' elif action == 'finv': h = solve(f == var('y'), x)[0].rhs() lbl = 'f^{-1}(y)' elif action == 'f+a': h = f+a lbl = 'f + a' elif action == 'f-a': h = f-a lbl = 'f - a' elif action == 'f*a': h = f*a lbl = 'f \\times a' elif action == 'f/a': h = f/a lbl = '\\frac{f}{a}' elif action == 'f^a': h = f^a lbl = 'f^a' elif action == 'f^a': h = f^a lbl = 'f^a' elif action == 'f(x+a)': h = f(x+a) lbl = 'f(x+a)' elif action == 'f(x*a)': h = f(x*a) lbl = 'f(xa)' elif action == 'f+g': h = f+g lbl = 'f + g' elif action == 'f-g': h = f-g lbl = 'f - g' elif action == 'f*g': h = f*g lbl = 'f \\times g' elif action == 'f/g': h = f/g lbl = '\\frac{f}{g}' elif action == 'f(g)': h = f(g) lbl = 'f(g)' html('<center><font color="red">$f = %s$</font></center>'%latex(f)) html('<center><font color="green">$g = %s$</font></center>'%latex(g)) html('<center><font color="blue"><b>$h = %s = %s$</b></font></center>'%(lbl, latex(h))) if do_plot: P = plot(f, xrange, color='red', thickness=2) + \ plot(g, xrange, color='green', thickness=2) + \ plot(h, xrange, color='blue', thickness=2) if yrange == 'auto': show(P, xmin=xrange[0], xmax=xrange[1]) else: yrange = sage_eval(yrange) show(P, xmin=xrange[0], xmax=xrange[1], ymin=yrange[0], ymax=yrange[1]) }}} attachment:funtool.png === Newton-Raphson Root Finding === by Neal Holtz This allows user to display the Newton-Raphson procedure one step at a time. It uses the heuristic that, if any of the values of the controls change, then the procedure should be re-started, else it should be continued. {{{ # ideas from 'A simple tangent line grapher' by Marshall Hampton # http://wiki.sagemath.org/interact State = Data = None # globals to allow incremental additions to graphics @interact def newtraph(f = input_box(default=8*sin(x)*exp(-x)-1, label='f(x)'), xmin = input_box(default=0), xmax = input_box(default=4*pi), x0 = input_box(default=3, label='x0'), show_calcs = ("Show Calcs",True), step = ['Next','Reset'] ): global State, Data prange = [xmin,xmax] state = [f,xmin,xmax,x0,show_calcs] if (state != State) or (step == 'Reset'): # when any of the controls change X = [RR(x0)] # restart the plot df = diff(f) Fplot = plot(f, prange[0], prange[1]) Data = [X, df, Fplot] State = state X, df, Fplot = Data i = len(X) - 1 # compute and append the next x value xi = X[i] fi = RR(f(xi)) fpi = RR(df(xi)) xip1 = xi - fi/fpi X.append(xip1) msg = xip1s = None # now check x value for reasonableness is_inf = False if abs(xip1) > 10E6*(xmax-xmin): is_inf = True show_calcs = True msg = 'Derivative is 0!' xip1s = latex(xip1.sign()*infinity) X.pop() elif not ((xmin - 0.5*(xmax-xmin)) <= xip1 <= (xmax + 0.5*(xmax-xmin))): show_calcs = True msg = 'x value out of range; probable divergence!' if xip1s is None: xip1s = '%.4g' % (xip1,) def Disp( s, color="blue" ): if show_calcs: html( """<font color="%s">$ %s $</font>""" % (color,s,) ) Disp( """f(x) = %s""" % (latex(f),) + """~~~~f'(x) = %s""" % (latex(df),) ) Disp( """i = %d""" % (i,) + """~~~~x_{%d} = %.4g""" % (i,xi) + """~~~~f(x_{%d}) = %.4g""" % (i,fi) + """~~~~f'(x_{%d}) = %.4g""" % (i,fpi) ) if msg: html( """<font color="red"><b>%s</b></font>""" % (msg,) ) c = "red" else: c = "blue" Disp( r"""x_{%d} = %.4g - ({%.4g})/({%.4g}) = %s""" % (i+1,xi,fi,fpi,xip1s), color=c ) Fplot += line( [(xi,0),(xi,fi)], linestyle=':', rgbcolor=(1,0,0) ) # vert dotted line Fplot += points( [(xi,0),(xi,fi)], rgbcolor=(1,0,0) ) labi = text( '\nx%d\n' % (i,), (xi,0), rgbcolor=(1,0,0), vertical_alignment="bottom" if fi < 0 else "top" ) if is_inf: xl = xi - 0.05*(xmax-xmin) xr = xi + 0.05*(xmax-xmin) yl = yr = fi else: xl = min(xi,xip1) - 0.02*(xmax-xmin) xr = max(xi,xip1) + 0.02*(xmax-xmin) yl = -(xip1-xl)*fpi yr = (xr-xip1)*fpi Fplot += points( [(xip1,0)], rgbcolor=(0,0,1) ) # new x value labi += text( '\nx%d\n' % (i+1,), (xip1,0), rgbcolor=(1,0,0), vertical_alignment="bottom" if fi < 0 else "top" ) Fplot += line( [(xl,yl),(xr,yr)], rgbcolor=(1,0,0) ) # tangent show( Fplot+labi, xmin = prange[0], xmax = prange[1] ) Data = [X, df, Fplot] }}} attachment:newtraph.png == Differential Equations == === Euler's Method in one variable === by Marshall Hampton. This needs some polishing but its usable as is. {{{ def tab_list(y, headers = None): ''' Converts a list into an html table with borders. ''' s = '<table border = 1>' if headers: for q in headers: s = s + '<th>' + str(q) + '</th>' for x in y: s = s + '<tr>' for q in x: s = s + '<td>' + str(q) + '</td>' s = s + '</tr>' s = s + '</table>' return s var('x y') @interact def euler_method(y_exact_in = input_box('-cos(x)+1.0', type = str, label = 'Exact solution = '), y_prime_in = input_box('sin(x)', type = str, label = "y' = "), start = input_box(0.0, label = 'x starting value: '), stop = input_box(6.0, label = 'x stopping value: '), startval = input_box(0.0, label = 'y starting value: '), nsteps = slider([2^m for m in range(0,10)], default = 10, label = 'Number of steps: '), show_steps = slider([2^m for m in range(0,10)], default = 8, label = 'Number of steps shown in table: ')): y_exact = lambda x: eval(y_exact_in) y_prime = lambda x,y: eval(y_prime_in) stepsize = float((stop-start)/nsteps) steps_shown = max(nsteps,show_steps) sol = [startval] xvals = [start] for step in range(nsteps): sol.append(sol[-1] + stepsize*y_prime(xvals[-1],sol[-1])) xvals.append(xvals[-1] + stepsize) sol_max = max(sol + [find_maximum_on_interval(y_exact,start,stop)[0]]) sol_min = min(sol + [find_minimum_on_interval(y_exact,start,stop)[0]]) show(plot(y_exact(x),start,stop,rgbcolor=(1,0,0))+line([[xvals[index],sol[index]] for index in range(len(sol))]),xmin=start,xmax = stop, ymax = sol_max, ymin = sol_min) if nsteps < steps_shown: table_range = range(len(sol)) else: table_range = range(0,floor(steps_shown/2)) + range(len(sol)-floor(steps_shown/2),len(sol)) html(tab_list([[i,xvals[i],sol[i]] for i in table_range], headers = ['step','x','y'])) }}} attachment:eulermethod.png == Linear Algebra == === Numerical instability of the classical Gram-Schmidt algorithm === by Marshall Hampton (tested by William Stein, who thinks this is really nice!) {{{ def GS_classic(a_list): ''' Given a list of vectors or a matrix, returns the QR factorization using the classical (and numerically unstable) Gram-Schmidt algorithm. ''' if type(a_list) != list: cols = a_list.cols() a_list = [x for x in cols] indices = range(len(a_list)) q = [] r = [[0 for i in indices] for j in indices] v = [a_list[i].copy() for i in indices] for i in indices: for j in range(0,i): r[j][i] = q[j].inner_product(a_list[i]) v[i] = v[i] - r[j][i]*q[j] r[i][i] = (v[i]*v[i])^(1/2) q.append(v[i]/r[i][i]) q = matrix([q[i] for i in indices]).transpose() return q, matrix(r) def GS_modern(a_list): ''' Given a list of vectors or a matrix, returns the QR factorization using the 'modern' Gram-Schmidt algorithm. ''' if type(a_list) != list: cols = a_list.cols() a_list = [x for x in cols] indices = range(len(a_list)) q = [] r = [[0 for i in indices] for j in indices] v = [a_list[i].copy() for i in indices] for i in indices: r[i][i] = v[i].norm(2) q.append(v[i]/r[i][i]) for j in range(i+1, len(indices)): r[i][j] = q[i].inner_product(v[j]) v[j] = v[j] - r[i][j]*q[i] q = matrix([q[i] for i in indices]).transpose() return q, matrix(r) html('<h2>Numerical instability of the classical Gram-Schmidt algorithm</h2>') @interact def gstest(precision = slider(range(3,53), default = 10), a1 = input_box([1,1/1000,1/1000]), a2 = input_box([1,1/1000,0]), a3 = input_box([1,0,1/1000])): myR = RealField(precision) displayR = RealField(5) html('precision in bits: ' + str(precision) + '<br>') A = matrix([a1,a2,a3]) A = [vector(myR,x) for x in A] qn, rn = GS_classic(A) qb, rb = GS_modern(A) html('Classical Gram-Schmidt:') show(matrix(displayR,qn)) html('Stable Gram-Schmidt:') show(matrix(displayR,qb)) }}} attachment:GramSchmidt.png === Linear transformations === by Jason Grout A square matrix defines a linear transformation which rotates and/or scales vectors. In the interact command below, the red vector represents the original vector (v) and the blue vector represents the image w under the linear transformation. You can change the angle and length of v by changing theta and r. {{{ @interact def linear_transformation(theta=slider(0, 2*pi, .1), r=slider(0.1, 2, .1, default=1)): A=matrix([[1,-1],[-1,1/2]]) v=vector([r*cos(theta), r*sin(theta)]) w = A*v circles = sum([circle((0,0), radius=i, rgbcolor=(0,0,0)) for i in [1..2]]) print jsmath("v = %s,\; %s v=%s"%(v.n(4),latex(A),w.n(4))) show(v.plot(rgbcolor=(1,0,0))+w.plot(rgbcolor=(0,0,1))+circles,aspect_ratio=1) }}} attachment:Linear-Transformations.png === Singular value decomposition === by Marshall Hampton {{{ import scipy.linalg as lin var('t') def rotell(sig,umat,t,offset=0): temp = matrix(umat)*matrix(2,1,[sig[0]*cos(t),sig[1]*sin(t)]) return [offset+temp[0][0],temp[1][0]] @interact def svd_vis(a11=slider(-1,1,.05,1),a12=slider(-1,1,.05,1),a21=slider(-1,1,.05,0),a22=slider(-1,1,.05,1),ofs= selector(['Off','On'],label='offset image from domain')): rf_low = RealField(12) my_mat = matrix(rf_low,2,2,[a11,a12,a21,a22]) u,s,vh = lin.svd(my_mat.numpy()) if ofs == 'On': offset = 3 fsize = 6 colors = [(1,0,0),(0,0,1),(1,0,0),(0,0,1)] else: offset = 0 fsize = 5 colors = [(1,0,0),(0,0,1),(.7,.2,0),(0,.3,.7)] vvects = sum([arrow([0,0],matrix(vh).row(i),rgbcolor = colors[i]) for i in (0,1)]) uvects = Graphics() for i in (0,1): if s[i] != 0: uvects += arrow([offset,0],vector([offset,0])+matrix(s*u).column(i),rgbcolor = colors[i+2]) html('<h3>Singular value decomposition: image of the unit circle and the singular vectors</h3>') print jsmath("A = %s = %s %s %s"%(latex(my_mat), latex(matrix(rf_low,u.tolist())), latex(matrix(rf_low,2,2,[s[0],0,0,s[1]])), latex(matrix(rf_low,vh.tolist())))) image_ell = parametric_plot(rotell(s,u,t, offset),0,2*pi) graph_stuff=circle((0,0),1)+image_ell+vvects+uvects graph_stuff.set_aspect_ratio(1) show(graph_stuff,frame = False,axes=False,figsize=[fsize,fsize]) }}} attachment:svd1.png === Discrete Fourier Transform === by Marshall Hampton {{{ import scipy.fftpack as Fourier @interact def discrete_fourier(f = input_box(default=sum([sin(k*x) for k in range(1,5,2)])), scale = slider(.1,20,.1,5)): var('x') pbegin = -float(pi)*scale pend = float(pi)*scale html("<h3>Function plot and its discrete Fourier transform</h3>") show(plot(f, pbegin, pend, plot_points = 512), figsize = [4,3]) f_vals = [f(ind) for ind in srange(pbegin, pend,(pend-pbegin)/512.0)] my_fft = Fourier.fft(f_vals) show(list_plot([abs(x) for x in my_fft], plotjoined=True), figsize = [4,3]) }}} attachment:dfft1.png == Algebra == === Groebner fan of an ideal === by Marshall Hampton; (needs sage-2.11 or higher, with gfan-0.3 interface) {{{ @interact def gfan_browse(p1 = input_box('x^3+y^2',type = str, label='polynomial 1: '), p2 = input_box('y^3+z^2',type = str, label='polynomial 2: '), p3 = input_box('z^3+x^2',type = str, label='polynomial 3: ')): R.<x,y,z> = PolynomialRing(QQ,3) i1 = ideal(R(p1),R(p2),R(p3)) gf1 = i1.groebner_fan() testr = gf1.render() html('Groebner fan of the ideal generated by: ' + str(p1) + ', ' + str(p2) + ', ' + str(p3)) show(testr, axes = False, figsize=[8,8*(3^(.5))/2]) }}} attachment:gfan_interact.png == Number Theory == === Continued Fraction Plotter === by William Stein {{{ @interact def _(number=e, ymax=selector([None,5,20,..,400],nrows=2), clr=Color('purple'), prec=[500,1000,..,5000]): c = list(continued_fraction(RealField(prec)(number))); print c show(line([(i,z) for i, z in enumerate(c)],rgbcolor=clr),ymax=ymax,figsize=[10,2]) }}} attachment:contfracplot.png === Illustrating the prime number thoerem === by William Stein {{{ @interact def _(N=(100,(2..2000))): html("<font color='red'>$\pi(x)$</font> and <font color='blue'>$x/(\log(x)-1)$</font> for $x < %s$"%N) show(plot(prime_pi, 0, N, rgbcolor='red') + plot(x/(log(x)-1), 5, N, rgbcolor='blue')) }}} attachment:primes.png === Computing Generalized Bernoulli Numbers === by William Stein (Sage-2.10.3) {{{ @interact def _(m=selector([1..15],nrows=2), n=(7,(3..10))): G = DirichletGroup(m) s = "<h3>First n=%s Bernoulli numbers attached to characters with modulus m=%s</h3>"%(n,m) s += '<table border=1>' s += '<tr bgcolor="#edcc9c"><td align=center>$\\chi$</td><td>Conductor</td>' + \ ''.join('<td>$B_{%s,\chi}$</td>'%k for k in [1..n]) + '</tr>' for eps in G.list(): v = ''.join(['<td align=center bgcolor="#efe5cd">$%s$</td>'%latex(eps.bernoulli(k)) for k in [1..n]]) s += '<tr><td bgcolor="#edcc9c">%s</td><td bgcolor="#efe5cd" align=center>%s</td>%s</tr>\n'%( eps, eps.conductor(), v) s += '</table>' html(s) }}} attachment:bernoulli.png === Fundamental Domains of SL_2(ZZ) === by Robert Miller {{{ L = [[-0.5, 2.0^(x/100.0) - 1 + sqrt(3.0)/2] for x in xrange(1000, -1, -1)] R = [[0.5, 2.0^(x/100.0) - 1 + sqrt(3.0)/2] for x in xrange(1000)] xes = [x/1000.0 for x in xrange(-500,501,1)] M = [[x,abs(sqrt(x^2-1))] for x in xes] fundamental_domain = L+M+R fundamental_domain = [[x-1,y] for x,y in fundamental_domain] @interact def _(gen = selector(['t+1', 't-1', '-1/t'], nrows=1)): global fundamental_domain if gen == 't+1': fundamental_domain = [[x+1,y] for x,y in fundamental_domain] elif gen == 't-1': fundamental_domain = [[x-1,y] for x,y in fundamental_domain] elif gen == '-1/t': new_dom = [] for x,y in fundamental_domain: sq_mod = x^2 + y^2 new_dom.append([(-1)*x/sq_mod, y/sq_mod]) fundamental_domain = new_dom P = polygon(fundamental_domain) P.ymax(1.2); P.ymin(-0.1) P.show() }}} attachment:fund_domain.png === Computing modular forms === by William Stein {{{ j = 0 @interact def _(N=[1..100], k=selector([2,4,..,12],nrows=1), prec=(3..40), group=[(Gamma0, 'Gamma0'), (Gamma1, 'Gamma1')]): M = CuspForms(group(N),k) print j; global j; j += 1 print M; print '\n'*3 print "Computing basis...\n\n" if M.dimension() == 0: print "Space has dimension 0" else: prec = max(prec, M.dimension()+1) for f in M.basis(): view(f.q_expansion(prec)) print "\n\n\nDone computing basis." }}} attachment:modformbasis.png === Computing the cuspidal subgroup === by William Stein {{{ html('<h1>Cuspidal Subgroups of Modular Jacobians J0(N)</h1>') @interact def _(N=selector([1..8*13], ncols=8, width=10, default=10)): A = J0(N) print A.cuspidal_subgroup() }}} attachment:cuspgroup.png === A Charpoly and Hecke Operator Graph === by William Stein {{{ # Note -- in Sage-2.10.3; multiedges are missing in plots; loops are missing in 3d plots @interact def f(N = prime_range(11,400), p = selector(prime_range(2,12),nrows=1), three_d = ("Three Dimensional", False)): S = SupersingularModule(N) T = S.hecke_matrix(p) G = Graph(T, multiedges=True, loops=not three_d) html("<h1>Charpoly and Hecke Graph: Level %s, T_%s</h1>"%(N,p)) show(T.charpoly().factor()) if three_d: show(G.plot3d(), aspect_ratio=[1,1,1]) else: show(G.plot(),figsize=7) }}} attachment:heckegraph.png === Demonstrating the Diffie-Hellman Key Exchange Protocol === by Timothy Clemans (refereed by William Stein) {{{ @interact def diffie_hellman(button=selector(["New example"],label='',buttons=True), bits=("Number of bits of prime", (8,12,..512))): maxp = 2^bits p = random_prime(maxp) k = GF(p) if bits>100: g = k(2) else: g = k.multiplicative_generator() a = ZZ.random_element(10, maxp) b = ZZ.random_element(10, maxp) print """ <html> <style> .gamodp { background:yellow } .gbmodp { background:orange } .dhsame { color:green; font-weight:bold } </style> <h2>%s-Bit Diffie-Hellman Key Exchange</h2> <ol style="color:#000;font:12px Arial, Helvetica, sans-serif"> <li>Alice and Bob agree to use the prime number p=%s and base g=%s.</li> <li>Alice chooses the secret integer a=%s, then sends Bob (<span class="gamodp">g<sup>a</sup> mod p</span>):<br/>%s<sup>%s</sup> mod %s = <span class="gamodp">%s</span>.</li> <li>Bob chooses the secret integer b=%s, then sends Alice (<span class="gbmodp">g<sup>b</sup> mod p</span>):<br/>%s<sup>%s</sup> mod %s = <span class="gbmodp">%s</span>.</li> <li>Alice computes (<span class="gbmodp">g<sup>b</sup> mod p</span>)<sup>a</sup> mod p:<br/>%s<sup>%s</sup> mod %s = <span class="dhsame">%s</span>.</li> <li>Bob computes (<span class="gamodp">g<sup>a</sup> mod p</span>)<sup>b</sup> mod p:<br/>%s<sup>%s</sup> mod %s = <span class="dhsame">%s</span>.</li> </ol></html> """ % (bits, p, g, a, g, a, p, (g^a), b, g, b, p, (g^b), (g^b), a, p, (g^ b)^a, g^a, b, p, (g^a)^b) }}} attachment:dh.png === Plotting an elliptic curve over a finite field === {{{ E = EllipticCurve('37a') @interact def _(p=slider(prime_range(1000), default=389)): show(E) print "p = %s"%p show(E.change_ring(GF(p)).plot(),xmin=0,ymin=0) }}} attachment:ellffplot.png == Web applications == === CO2 data plot, fetched from NOAA === by Marshall Hampton {{{ import urllib2 as U @interact def mauna_loa_co2(start_date = slider(1958,2010,1,1958), end_date = slider(1958, 2010,1,2009), Update = selector(['Update'], buttons=True, label = '')): co2data = U.urlopen('ftp://ftp.cmdl.noaa.gov/ccg/co2/trends/co2_mm_mlo.txt').readlines() datalines = [] for a_line in co2data: if a_line.find('Creation:') != -1: cdate = a_line if a_line[0] != '#': temp = a_line.replace('\n','').split(' ') temp = [float(q) for q in temp if q != ''] datalines.append(temp) html('<h3>CO2 monthly averages at Mauna Loa (interpolated), from NOAA/ESRL data</h3>') html('<h4>'+cdate+'</h4>') sel_data = [[q[2],q[4]] for q in datalines if start_date < q[2] < end_date] c_max = max([q[1] for q in sel_data]) c_min = min([q[1] for q in sel_data]) show(list_plot([[q[2],q[4]] for q in datalines], plotjoined=True, rgbcolor=(1,0,0)), xmin = start_date, ymin = c_min-2, axes = True, xmax = end_date, ymax = c_max+3, frame = False) }}} attachment:mauna_loa_co2.png == Bioinformatics == === Web app: protein browser === by Marshall Hampton (tested by William Stein) {{{ import urllib2 as U @interact def protein_browser(GenBank_ID = input_box('165940577', type = str), file_type = selector([(1,'fasta'),(2,'GenPept')])): if file_type == 2: gen_str = 'http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&sendto=t&id=' else: gen_str = 'http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=protein&sendto=t&dopt=fasta&id=' f = U.urlopen(gen_str + GenBank_ID) g = f.read() f.close() html(g) }}} attachment:biobrowse.png === Coalescent simulator === by Marshall Hampton {{{ def next_gen(x, selection=1.0): '''Creates the next generation from the previous; also returns parent-child indexing list''' next_x = [] for ind in range(len(x)): if random() < (1 + selection)/len(x): rind = 0 else: rind = int(round(random()*(len(x)-1)+1/2)) next_x.append((x[rind],rind)) next_x.sort() return [[x[0] for x in next_x],[x[1] for x in next_x]] def coal_plot(some_data): '''Creates a graphics object from coalescent data''' gens = some_data[0] inds = some_data[1] gen_lines = line([[0,0]]) pts = Graphics() ngens = len(gens) gen_size = len(gens[0]) for x in range(gen_size): pts += point((x,ngens-1), hue = gens[0][x]/float(gen_size*1.1)) p_frame = line([[-.5,-.5],[-.5,ngens-.5], [gen_size-.5,ngens-.5], [gen_size-.5,-.5], [-.5,-.5]]) for g in range(1,ngens): for x in range(gen_size): old_x = inds[g-1][x] gen_lines += line([[x,ngens-g-1],[old_x,ngens-g]], hue = gens[g-1][old_x]/float(gen_size*1.1)) pts += point((x,ngens-g-1), hue = gens[g][x]/float(gen_size*1.1)) return pts+gen_lines+p_frame d_field = RealField(10) @interact def coalescents(pop_size = slider(2,100,1,15,'Population size'), selection = slider(-1,1,.1,0, 'Selection for first taxon'), s = selector(['Again!'], label='Refresh', buttons=True)): print 'Population size: ' + str(pop_size) print 'Selection coefficient for first taxon: ' + str(d_field(selection)) start = [i for i in range(pop_size)] gens = [start] inds = [] while gens[-1][0] != gens[-1][-1]: g_index = len(gens) - 1 n_gen = next_gen(gens[g_index], selection = selection) gens.append(n_gen[0]) inds.append(n_gen[1]) coal_data1 = [gens,inds] print 'Generations until coalescence: ' + str(len(gens)) show(coal_plot(coal_data1), axes = False, figsize = [8,4.0*len(gens)/pop_size], ymax = len(gens)-1) }}} attachment:coalescent.png == Miscellaneous Graphics == === Catalog of 3D Parametric Plots === {{{ var('u,v') plots = ['Two Interlinked Tori', 'Star of David', 'Double Heart', 'Heart', 'Green bowtie', "Boy's Surface", "Maeder's Owl", 'Cross cap'] plots.sort() @interact def _(example=selector(plots, buttons=True, nrows=2), tachyon=("Raytrace", False), frame = ('Frame', False), opacity=(1,(0.1,1))): url = '' if example == 'Two Interlinked Tori': f1 = (4+(3+cos(v))*sin(u), 4+(3+cos(v))*cos(u), 4+sin(v)) f2 = (8+(3+cos(v))*cos(u), 3+sin(v), 4+(3+cos(v))*sin(u)) p1 = parametric_plot3d(f1, (u,0,2*pi), (v,0,2*pi), color="red", opacity=opacity) p2 = parametric_plot3d(f2, (u,0,2*pi), (v,0,2*pi), color="blue",opacity=opacity) P = p1 + p2 elif example == 'Star of David': f_x = cos(u)*cos(v)*(abs(cos(3*v/4))^500 + abs(sin(3*v/4))^500)^(-1/260)*(abs(cos(4*u/4))^200 + abs(sin(4*u/4))^200)^(-1/200) f_y = cos(u)*sin(v)*(abs(cos(3*v/4))^500 + abs(sin(3*v/4))^500)^(-1/260)*(abs(cos(4*u/4))^200 + abs(sin(4*u/4))^200)^(-1/200) f_z = sin(u)*(abs(cos(4*u/4))^200 + abs(sin(4*u/4))^200)^(-1/200) P = parametric_plot3d([f_x, f_y, f_z], (u, -pi, pi), (v, 0, 2*pi),opacity=opacity) elif example == 'Double Heart': f_x = ( abs(v) - abs(u) - abs(tanh((1/sqrt(2))*u)/(1/sqrt(2))) + abs(tanh((1/sqrt(2))*v)/(1/sqrt(2))) )*sin(v) f_y = ( abs(v) - abs(u) - abs(tanh((1/sqrt(2))*u)/(1/sqrt(2))) - abs(tanh((1/sqrt(2))*v)/(1/sqrt(2))) )*cos(v) f_z = sin(u)*(abs(cos(4*u/4))^1 + abs(sin(4*u/4))^1)^(-1/1) P = parametric_plot3d([f_x, f_y, f_z], (u, 0, pi), (v, -pi, pi),opacity=opacity) elif example == 'Heart': f_x = cos(u)*(4*sqrt(1-v^2)*sin(abs(u))^abs(u)) f_y = sin(u) *(4*sqrt(1-v^2)*sin(abs(u))^abs(u)) f_z = v P = parametric_plot3d([f_x, f_y, f_z], (u, -pi, pi), (v, -1, 1), frame=False, color="red",opacity=opacity) elif example == 'Green bowtie': f_x = sin(u) / (sqrt(2) + sin(v)) f_y = sin(u) / (sqrt(2) + cos(v)) f_z = cos(u) / (1 + sqrt(2)) P = parametric_plot3d([f_x, f_y, f_z], (u, -pi, pi), (v, -pi, pi), frame=False, color="green",opacity=opacity) elif example == "Boy's Surface": url = "http://en.wikipedia.org/wiki/Boy's_surface" fx = 2/3* (cos(u)* cos(2*v) + sqrt(2)* sin(u)* cos(v))* cos(u) / (sqrt(2) - sin(2*u)* sin(3*v)) fy = 2/3* (cos(u)* sin(2*v) - sqrt(2)* sin(u)* sin(v))* cos(u) / (sqrt(2) - sin(2*u)* sin(3*v)) fz = sqrt(2)* cos(u)* cos(u) / (sqrt(2) - sin(2*u)* sin(3*v)) P = parametric_plot3d([fx, fy, fz], (u, -2*pi, 2*pi), (v, 0, pi), plot_points = [90,90], frame=False, color="orange",opacity=opacity) elif example == "Maeder's Owl": fx = v *cos(u) - 0.5* v^2 * cos(2* u) fy = -v *sin(u) - 0.5* v^2 * sin(2* u) fz = 4 *v^1.5 * cos(3 *u / 2) / 3 P = parametric_plot3d([fx, fy, fz], (u, -2*pi, 2*pi), (v, 0, 1),plot_points = [90,90], frame=False, color="purple",opacity=opacity) elif example =='Cross cap': url = 'http://en.wikipedia.org/wiki/Cross-cap' fx = (1+cos(v))*cos(u) fy = (1+cos(v))*sin(u) fz = -tanh((2/3)*(u-pi))*sin(v) P = parametric_plot3d([fx, fy, fz], (u, 0, 2*pi), (v, 0, 2*pi), frame=False, color="red",opacity=opacity) else: print "Bug selecting plot?" return html('<h2>%s</h2>'%example) if url: html('<h3><a target="_new" href="%s">%s</a></h3>'%(url,url)) show(P, viewer='tachyon' if tachyon else 'jmol', frame=frame) }}} attachment:parametricplot3d.png === Interactive rotatable raytracing with Tachyon3d === {{{ C = cube(color=['red', 'green', 'blue'], aspect_ratio=[1,1,1], viewer='tachyon') + sphere((1,0,0),0.2) @interact def example(theta=(0,2*pi), phi=(0,2*pi), zoom=(1,(1,4))): show(C.rotate((0,0,1), theta).rotate((0,1,0),phi), zoom=zoom) }}} attachment:tachyonrotate.png === Interactive 3d plotting === {{{ var('x,y') @interact def example(clr=Color('orange'), f=4*x*exp(-x^2-y^2), xrange='(-2, 2)', yrange='(-2,2)', zrot=(0,pi), xrot=(0,pi), zoom=(1,(1/2,3)), square_aspect=('Square Frame', False), tachyon=('Ray Tracer', True)): xmin, xmax = sage_eval(xrange); ymin, ymax = sage_eval(yrange) P = plot3d(f, (x, xmin, xmax), (y, ymin, ymax), color=clr) html('<h1>Plot of $f(x,y) = %s$</h1>'%latex(f)) aspect_ratio = [1,1,1] if square_aspect else [1,1,1/2] show(P.rotate((0,0,1), -zrot).rotate((1,0,0),xrot), viewer='tachyon' if tachyon else 'jmol', figsize=6, zoom=zoom, frame=False, frame_aspect_ratio=aspect_ratio) }}} attachment:tachyonplot3d.png [[Anchor(eggpaint)]] === Somewhat Silly Egg Painter === by Marshall Hampton (refereed by William Stein) {{{ var('s,t') g(s) = ((0.57496*sqrt(121 - 16.0*s^2))/sqrt(10.+ s)) def P(color, rng): return parametric_plot3d((cos(t)*g(s), sin(t)*g(s), s), (s,rng[0],rng[1]), (t,0,2*pi), plot_points = [150,150], rgbcolor=color, frame = False, opacity = 1) colorlist = ['red','blue','red','blue'] @interact def _(band_number = selector(range(1,5)), current_color = Color('red')): html('<h1 align=center>Egg Painter</h1>') colorlist[band_number-1] = current_color egg = sum([P(colorlist[i],[-2.75+5.5*(i/4),-2.75+5.5*(i+1)/4]) for i in range(4)]) show(egg) }}} attachment:eggpaint.png |
{{attachment:taylor_series_animated.gif}} |
Sage Interactions
This is a collection of pages demonstrating the use of the interact command in Sage. It should be easy to just scroll through and copy/paste examples into sage notebooks. If you have suggestions on how to improve interact, add them here or email [email protected] . Of course, your own examples are also welcome!
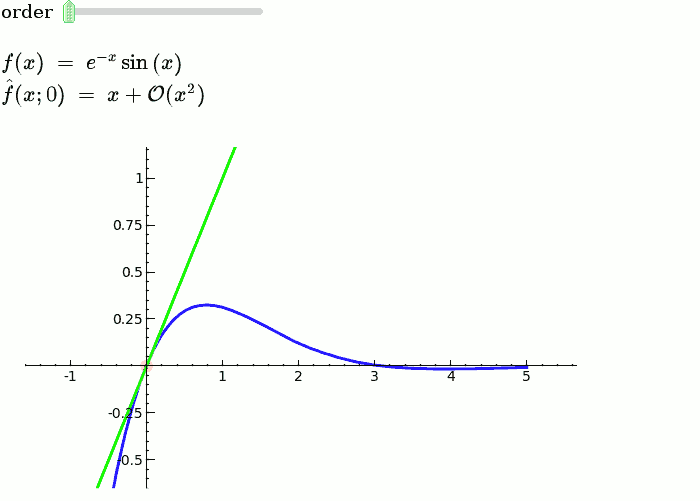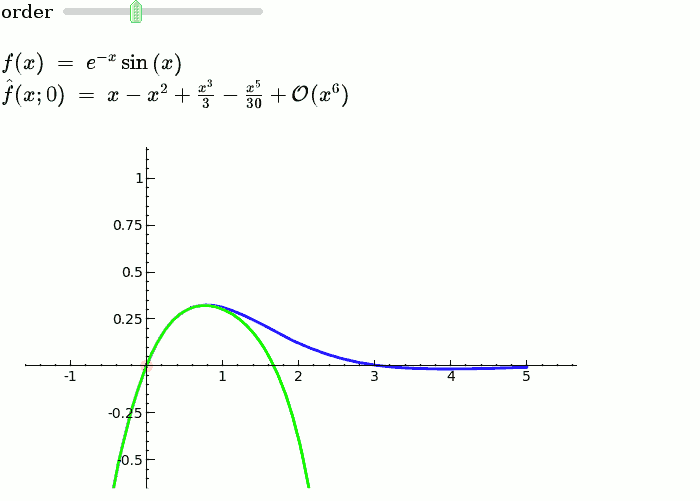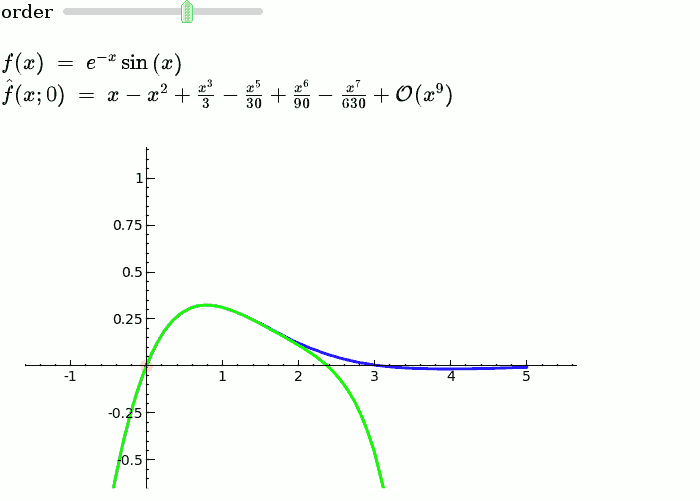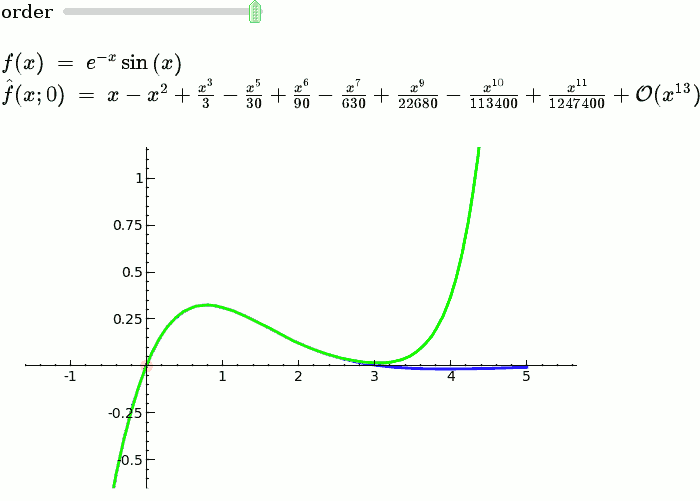
Explanatory example: Taylor Series
This is the code and a mockup animation of the interact command. It defines a slider, seen on top, that can be dragged. Once dragged, it changes the value of the variable "order" and the whole block of code gets evaluated. This principle can be seen in various examples presented on the pages above!
var('x')
x0 = 0
f = sin(x)*e^(-x)
p = plot(f,-1,5, thickness=2)
dot = point((x0,f(x=x0)),pointsize=80,rgbcolor=(1,0,0))
@interact
def _(order=(1..12)):
ft = f.taylor(x,x0,order)
pt = plot(ft,-1, 5, color='green', thickness=2)
html('$f(x)\;=\;%s$'%latex(f))
html('$\hat{f}(x;%s)\;=\;%s+\mathcal{O}(x^{%s})$'%(x0,latex(ft),order+1))
show(dot + p + pt, ymin = -.5, ymax = 1)