|
Size: 2330
Comment:
|
Size: 5481
Comment:
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 2: | Line 2: |
| goto [:interact:interact main page] | goto [[interact|interact main page]] |
| Line 4: | Line 4: |
| [[TableOfContents]] == Automorphism Groups of some Graphs == by William Stein (I spent less than five minutes on this): {{{ @interact def _(graph=['CycleGraph', 'CubeGraph', 'RandomGNP'], n=selector([1..10],nrows=1), p=selector([10,20,..,100],nrows=1)): print graph if graph == 'CycleGraph': print "n (=%s): number of vertices"%n G = graphs.CycleGraph(n) elif graph == 'CubeGraph': if n > 8: print "n reduced to 8" n = 8 print "n (=%s): dimension"%n G = graphs.CubeGraph(n) elif graph == 'RandomGNP': print "n (=%s) vertices"%n print "p (=%s%%) probability"%p G = graphs.RandomGNP(n, p/100.0) print G.automorphism_group() show(plot(G)) }}} attachment:autograph.png |
<<TableOfContents>> |
| Line 35: | Line 8: |
| (Very simple so far.) {{{ |
{{{#!sagecell grs = ['BalancedTree', 'BullGraph', 'ChvatalGraph', 'CirculantGraph', 'CircularLadderGraph', 'ClawGraph', 'CompleteBipartiteGraph', 'CompleteGraph', 'CubeGraph', 'CycleGraph', 'DegreeSequence', 'DegreeSequenceConfigurationModel', 'DegreeSequenceExpected', 'DegreeSequenceTree', 'DesarguesGraph', 'DiamondGraph', 'DodecahedralGraph', 'DorogovtsevGoltsevMendesGraph', 'EmptyGraph', 'FlowerSnark', 'FruchtGraph', 'Grid2dGraph', 'GridGraph', 'HeawoodGraph', 'HexahedralGraph', 'HoffmanSingletonGraph', 'HouseGraph', 'HouseXGraph', 'IcosahedralGraph', 'KrackhardtKiteGraph', 'LCFGraph', 'LadderGraph', 'LollipopGraph', 'MoebiusKantorGraph', 'OctahedralGraph', 'PappusGraph', 'PathGraph', 'PetersenGraph', 'RandomBarabasiAlbert', 'RandomGNM', 'RandomGNP', 'RandomHolmeKim', 'RandomLobster', 'RandomNewmanWattsStrogatz', 'RandomRegular', 'RandomTreePowerlaw', 'StarGraph', 'TetrahedralGraph', 'ThomsenGraph', 'WheelGraph'] |
| Line 72: | Line 45: |
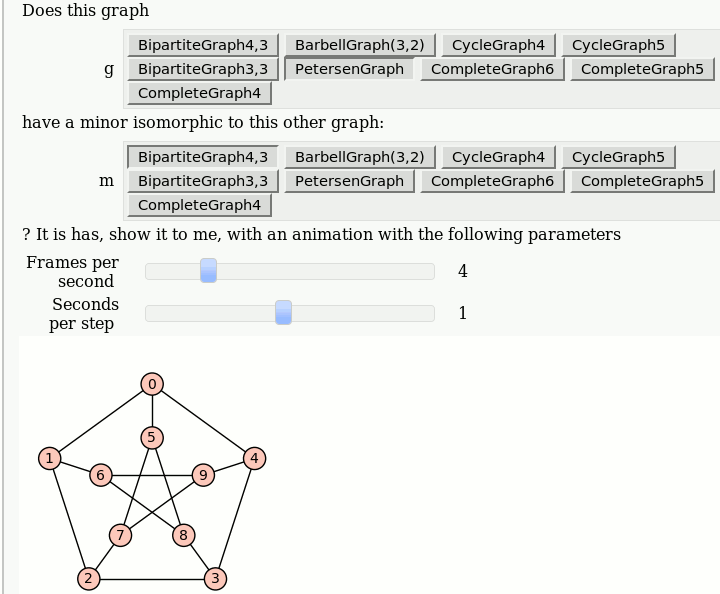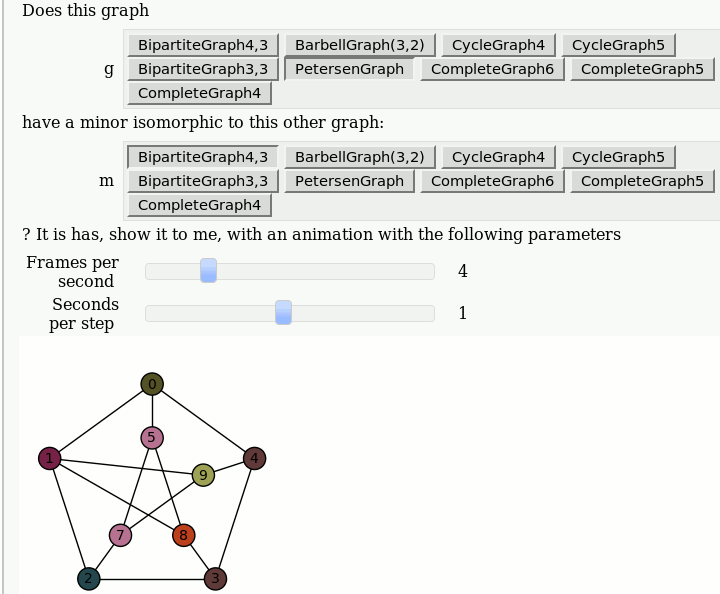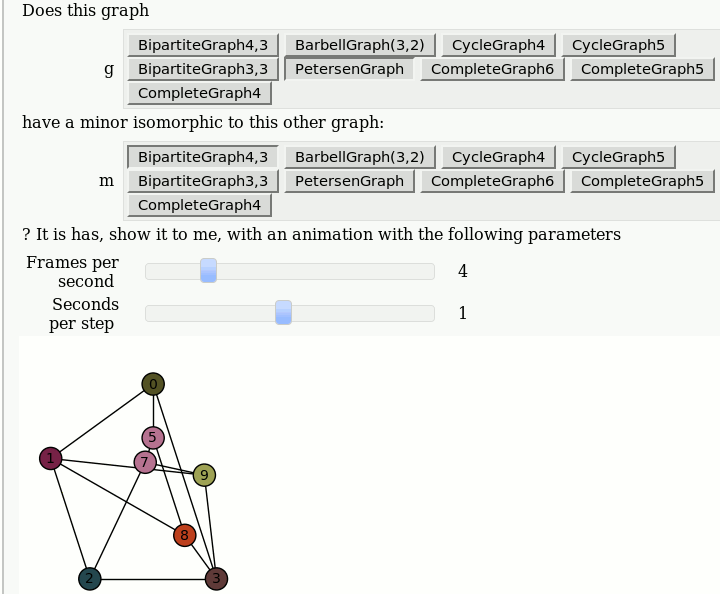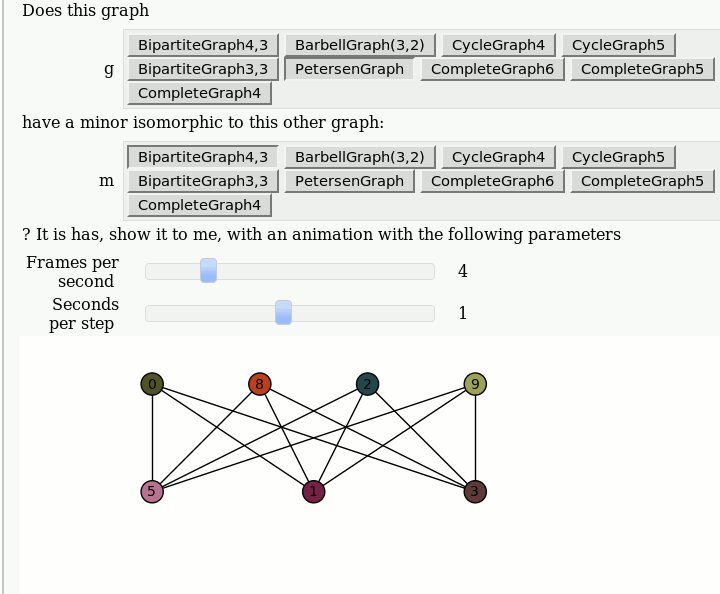
| attachment:graph_browse.png | {{attachment:graph_browse.png}} == Automorphism Groups of some Graphs == by William Stein: {{{#!sagecell @interact def _(graph=['CycleGraph', 'CubeGraph', 'RandomGNP'], n=selector([1..10],nrows=1), p=selector([10,20,..,100],nrows=1)): print graph if graph == 'CycleGraph': print "n = %s (number of vertices)"%n G = graphs.CycleGraph(n) elif graph == 'CubeGraph': if n > 8: print "n reduced to 8" n = 8 print "n = %s (dimension)"%n G = graphs.CubeGraph(n) elif graph == 'RandomGNP': print "n = %s (number of vertices)"%n print "p = %s%% (edge probability)"%p G = graphs.RandomGNP(n, p/100.0) print G.automorphism_group() show(plot(G)) }}} {{attachment:auto_graph2.png}} == View an induced subgraph == by Jason Grout {{{#!sagecell m=random_matrix(ZZ,10,density=.5) a=DiGraph(m) @interact def show_subgraph(to_delete=selector(range(10),buttons=True)): vertices=set(range(10)) subgraph_vertices=vertices.difference([to_delete]) html.table([["Original", "New"], [plot(a,save_pos=True), plot(a.subgraph(subgraph_vertices))]], header=True) }}} {{attachment:subgraph-interact.png}} == Animations of Graph Minors == by Pablo Angulo Note: you should use "Upload" or "Create a new file" to attach to the worksheet a file called wagner.sage with the following [[interact/graph_theory/wagner.sage| content]], and then you can use the code below: {{{#!sagecell attach(DATA + 'wagner.sage') graph_list = {'CompleteGraph4':graphs.CompleteGraph(4), 'CompleteGraph5':graphs.CompleteGraph(5), 'CompleteGraph6':graphs.CompleteGraph(6), 'BipartiteGraph3,3':graphs.CompleteBipartiteGraph(3,3), 'BipartiteGraph4,3':graphs.CompleteBipartiteGraph(4,3), 'PetersenGraph':graphs.PetersenGraph(), 'CycleGraph4':graphs.CycleGraph(4), 'CycleGraph5':graphs.CycleGraph(5), 'BarbellGraph(3,2)':graphs.BarbellGraph(3,2) } @interact def _(u1 = text_control(value='Does this graph'), g = selector(graph_list.keys(), buttons = True), u2 = text_control(value='have a minor isomorphic to this other graph:'), m = selector(graph_list.keys(), buttons = True), u3 = text_control(value='''? It is has, show it to me, with an animation with the following parameters'''), frames = slider(1,15,1,4, label = 'Frames per second'), step_time = slider(1/10,2,1/10,1, label = 'Seconds per step')): g = graph_list[g] m = graph_list[m] if g == m: html('<h3>Please select two different graphs</h3>') return try: a = animate_minor(g, m, frames = frames) a.show(delay=100*step_time/frames) except ValueError: html('''<h3>The first graph have <strong>NO</strong> minor isomorphic to the second</h3>''') }}} {{attachment:wagner.gif}} |
Sage Interactions - Graph Theory
goto interact main page
Contents
Graph Browser
by Marshall Hampton
Automorphism Groups of some Graphs
by William Stein:
View an induced subgraph
by Jason Grout
Animations of Graph Minors
by Pablo Angulo
Note: you should use "Upload" or "Create a new file" to attach to the worksheet a file called wagner.sage with the following content, and then you can use the code below: