Sage Interactions - Algebra
goto interact main page
Contents
Groebner fan of an ideal
by Marshall Hampton; (needs sage-2.11 or higher, with gfan-0.3 interface)
@interact
def gfan_browse(p1 = input_box('x^3+y^2',type = str, label='polynomial 1: '), p2 = input_box('y^3+z^2',type = str, label='polynomial 2: '), p3 = input_box('z^3+x^2',type = str, label='polynomial 3: ')):
R.<x,y,z> = PolynomialRing(QQ,3)
i1 = ideal(R(p1),R(p2),R(p3))
gf1 = i1.groebner_fan()
testr = gf1.render()
html('Groebner fan of the ideal generated by: ' + str(p1) + ', ' + str(p2) + ', ' + str(p3))
show(testr, axes = False, figsize=[8,8*(3^(.5))/2])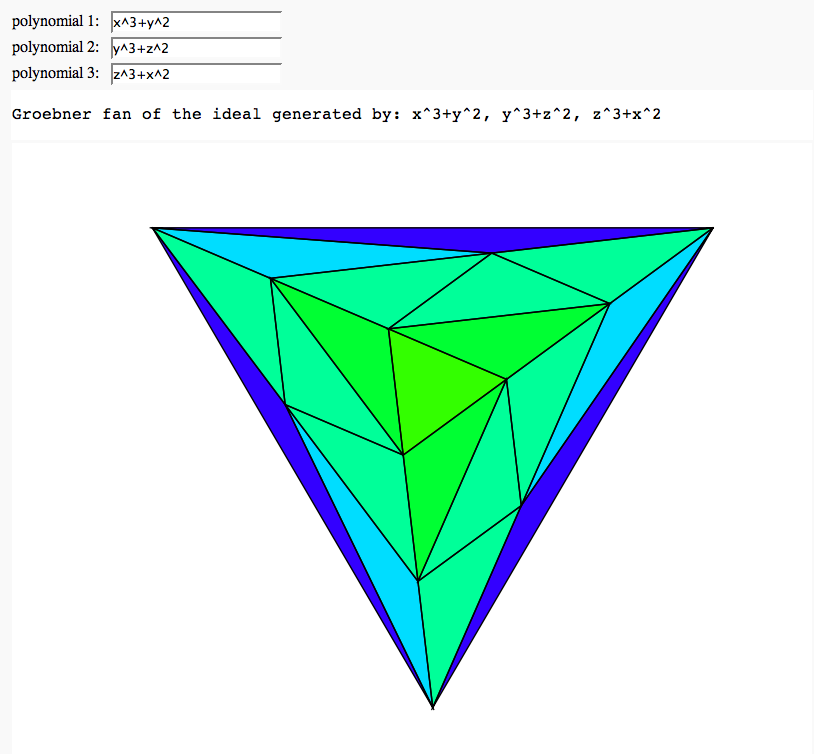
3D Groebner fan browser
by Marshall Hampton
def proj4_to_3(gfanobj, poly4):
fpoints = poly4.vertices()
tpoints = [gfanobj._embed_tetra(q) for q in fpoints]
adj_data = poly4.vertex_adjacencies()
edges = []
for adj in adj_data:
for vert in adj[1]:
if vert > adj[0]:
edges.append([tpoints[adj[0]],tpoints[vert]])
return edges, tpoints
from sage.plot.plot3d.index_face_set import IndexFaceSet
def render_solid(poly, color = 'blue', opacity = .5):
tri_faces = poly.triangulated_facial_incidences()
from sage.plot.plot3d.index_face_set import IndexFaceSet
return IndexFaceSet([q[1] for q in tri_faces], poly.vertices(), enclosed = True, color = color, opacity = opacity)
def render3d(a_gf, color_fan = True, verbose = False, highlights = 'all'):
g_cones = [q.groebner_cone() for q in a_gf.reduced_groebner_bases()]
g_cones_facets = [q.facets() for q in g_cones]
g_cones_ieqs = [a_gf._cone_to_ieq(q) for q in g_cones_facets]
# Now the cones are intersected with a plane:
cone_info = [ieq_to_vert(q,linearities=[[1,-1,-1,-1,-1]]) for q in g_cones_ieqs]
if verbose:
for x in cone_info:
print x.ieqs() + [[1,1,0,0,0],[1,0,1,0,0],[1,0,0,1,0],[1,0,0,0,1]]
print x.linearities()
print ""
cone_info = [Polyhedron(ieqs = x.ieqs() + [[1,1,0,0,0],[1,0,1,0,0],[1,0,0,1,0],[1,0,0,0,1]], linearities = x.linearities()) for x in cone_info]
if color_fan == True:
#using fixed color scheme
color_list = []
our_vars = list(a_gf.ring().gens())
degs = [[max([q.degree(avar) for q in b]) for avar in our_vars] for b in a_gf.reduced_groebner_bases()]
maxdegs = [max([float(q[i]) for q in degs]) for i in range(len(our_vars))]
color_list = [[b[0]/maxdegs[0],b[1]/maxdegs[1],(b[2]+b[3])/(maxdegs[2]+maxdegs[3])] for b in degs]
color_list = [tuple([c[i]/max(c) for i in range(3)]) for c in color_list]
faces = []
if highlights == 'all':
highlights = range(len(cone_info))
all_lines = []
i = 0
for cone_data in cone_info:
# cone_data is a Polyhedron.
try:
pdata = proj4_to_3(a_gf,cone_data)
cone_lines = pdata[0]
cone_verts = pdata[1]
if color_fan == True:
if i in highlights:
faces.append(render_solid(Polyhedron(vertices = cone_verts), color = color_list[i]))
i = i + 1
except:
print cone_data._rays
raise RuntimeError
for a_line in cone_lines:
all_lines.append(a_line)
if faces == []:
faceadds = Graphics()
else:
faceadds = sum(faces)
return sum([line3d(a_line) for a_line in all_lines]) + faceadds
R4.<w,x,y,z> = PolynomialRing(QQ,4)
temp_id = R4.ideal([w^3-x^2, x^3-y^21, y^3-w^2, z - x^2])
temp_gf4 = temp_id.groebner_fan()
temp_gf4_rbs = temp_gf4.reduced_groebner_bases()
gbdict = dict([['w^3-x^2, x^3-y^2, y^3-w^2, z - x^2',(temp_gf4,temp_gf4_rbs)]])
@interact
def Groebner_fan_browser(bsel = slider(0,100,.1,0,label='Individual basis selection', display_value = False), ideal_gens = input_box(default = 'w^3-x^2, x^3-y^2, y^3-w^2, z - x^2', type = str, label = "Ideal generators"), showall = checkbox(True, "Show me them all"), showbases = checkbox(False, "Show highlighted basis")):
html('<h3>Groebner fan 3D browser</h3> Enter 4 polynomials in the variables w,x,y,z<BR> <em>This may take forever if you are overambitious</em>')
R4.<w,x,y,z> = PolynomialRing(QQ,4)
if ideal_gens not in gbdict:
id_gens = R4.ideal(list(ideal_gens.split(',')))
print id_gens
gf4 = id_gens.groebner_fan()
gf4rbs = gf4.reduced_groebner_bases()
gbdict[ideal_gens] = (gf4,gf4rbs)
else:
gf4 = gbdict[ideal_gens][0]
gf4rbs = gbdict[ideal_gens][1]
bnumbers = len(gf4rbs)
b_select = [int(bsel*bnumbers/100.0)]
if showall: b_select = range(bnumbers)
if showbases:
for b in b_select:
show(gf4rbs[b])
show(render3d(gf4, highlights = b_select), frame = False)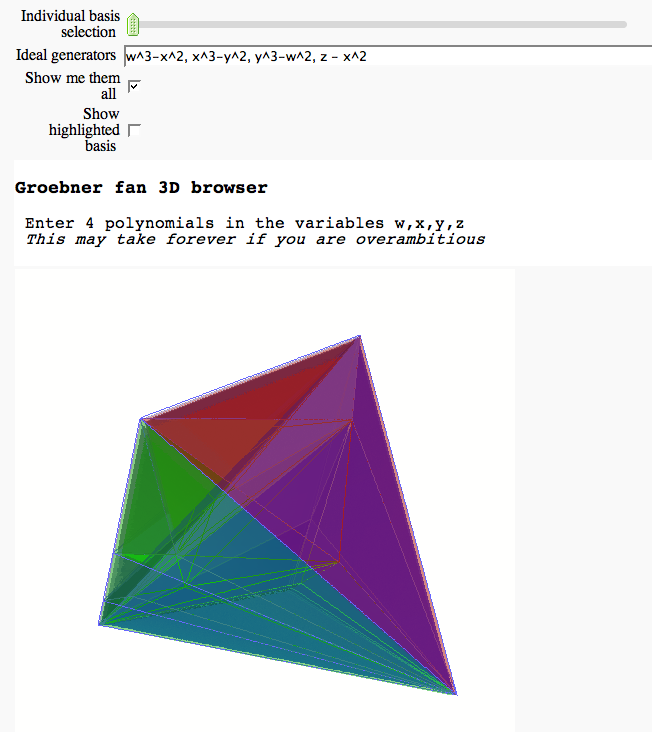
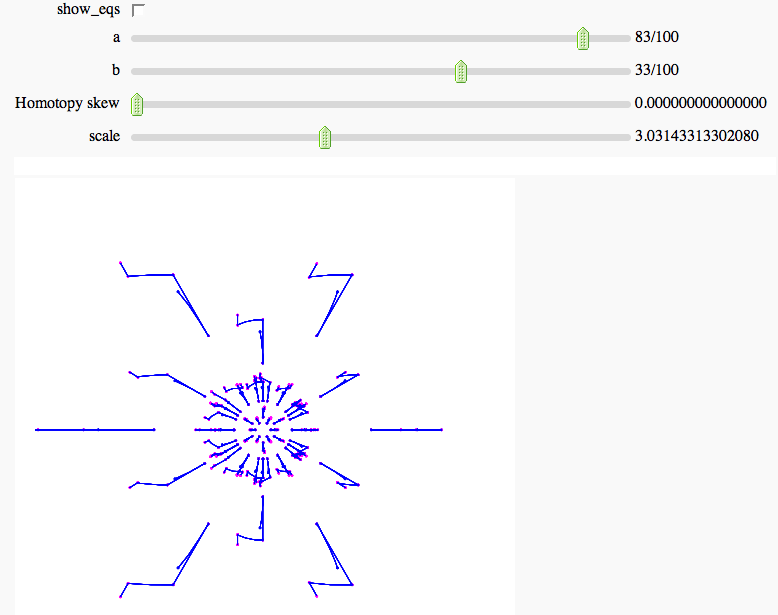
Numerical Solutions of Polynomial Systems with PHCpack
by Marshall Hampton; requires phcpack optional package (PHCpack written by Jan Verschelde). The example below is a two-parameter deformation of the cyclic-6 problem. Solution paths are tracked through the parameter homotopy.
from sage.interfaces.phc import phc
zringA.<z0,z1,z2,z3,z4,z5,a,b> = PolynomialRing(QQ,8)
cyclic6 = [z0 + z1 + z2 + z3 + z4 + z5+a,
z0*z1 + z1*z2 + z2*z3 + z3*z4 + z4*z5 + z5*z0,
z0*z1*z2 + z1*z2*z3 + z2*z3*z4 + z3*z4*z5 + z4*z5*z0 + z5*z0*z1,
z0*z1*z2*z3 + z1*z2*z3*z4 + z2*z3*z4*z5 + z3*z4*z5*z0 + z4*z5*z0*z1
+ z5*z0*z1*z2,
z0*z1*z2*z3*z4 + z1*z2*z3*z4*z5 + z2*z3*z4*z5*z0 + z3*z4*z5*z0*z1
+ z4*z5*z0*z1*z2 + z5*z0*z1*z2*z3,
z0*z1*z2*z3*z4*z5 - b]
zring.<z0,z1,z2,z3,z4,z5> = PolynomialRing(QQ,6)
z1 = [zring(x.subs({a:1/10, b:1/10})) for x in cyclic6]
s1 = phc.blackbox(z1,zring)
s1sas = s1.save_as_start(start_filename = DATA + 's1phc')
cstate = [open(DATA + 's1phc').read()]
def def_cyclic(ain, bin):
eqs = [zring(x.subs({a:ain, b:bin})) for x in cyclic6]
return eqs
slines2d = []
mpts = []
@interact
def tbp_tracker(show_eqs = checkbox(False),a = slider(-1,1,1/100,1/100), b = slider(-1,1,1/100,1/100), h_c_skew = slider(0,.1,.001,0.0, label='Homotopy skew'), scale = slider([2.0^x for x in srange(.1,4,.025)],default = 2^1.6)):
z_pt = phc._path_track_file(start_filename_or_string = cstate[-1], polys = def_cyclic(a,b), input_ring = zring, c_skew = h_c_skew)
cstate.append(open(z_pt).read())
z_pp = phc._parse_path_file(z_pt)
hue_v = len(cstate)/(len(cstate)+1)
znames = ['z0','z1','z2','z3','z4','z5']
for a_sol in z_pp:
for z in znames:
mpts.append(point([a_sol[0][z].real(), a_sol[0][z].imag()], hue=hue_v,pointsize=3))
mpts.append(point([a_sol[-1][z].real(), a_sol[-1][z].imag()], hue=hue_v,pointsize=3))
for a_sol in z_pp:
zlines = [[] for q in znames]
for data in a_sol:
for i in range(len(znames)):
zn = znames[i]
zlines[i].append([data[zn].real(), data[zn].imag()])
for zl in zlines:
slines2d.append(line(zl, thickness = .5))
show(sum(slines2d)+sum(mpts), figsize = [5,5], xmin = -scale, xmax=scale, ymin=-scale,ymax=scale, axes = false)
if show_eqs:
pols = def_cyclic(a,b)
for i in range(len(pols)):
show(pols[i])