|
Size: 4934
Comment:
|
Size: 43249
Comment: List of major features
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 4: | Line 4: |
| Sage 4.1.1 was released on August 14th, 2009. For the official, comprehensive release note, please refer to [[http://www.sagemath.org/src/announce/sage-4.1.1.txt|sage-4.1.1.txt]]. A nicely formatted version of this release tour can be found here. The following points are some of the foci of this release: * Improved data conversion between NumPy and Sage * Solaris support, Solaris specific bug fixes for NTL, zn_poly, Pari/GP, FLINT, MPFR, PolyBoRI, ATLAS * Upgrade/updates for about 8 standard packages * Three new optional packages: openopt, GLPK, p_group_cohomology |
|
| Line 7: | Line 15: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6510|#6510]] | * Adds method {{{__nonzero__()}}} to abelian groups (Taylor Sutton) [[http://trac.sagemath.org/sage_trac/ticket/6510|#6510]] --- New method {{{__nonzero__()}}} for the class {{{AbelianGroup_class}}} in {{{sage/groups/abelian_gps/abelian_group.py}}}. This method returns {{{True}}} if the abelian group in question is non-trivial: {{{#!python numbers=off sage: E = EllipticCurve([0, 82]) sage: T = E.torsion_subgroup() sage: bool(T) False sage: T.__nonzero__() False }}} |
| Line 13: | Line 29: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6159|#6159]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/2737|#2737]] |
* Implement {{{real_part()}}} and {{{imag_part()}}} for {{{CDF}}} and {{{CC}}} (Alex Ghitza) [[http://trac.sagemath.org/sage_trac/ticket/6159|#6159]] --- The name {{{real_part}}} is now an alias to the method {{{real()}}}; similarly, {{{imag_part}}} is now an alias to the method {{{imag()}}}. {{{#!python numbers=off sage: a = CDF(3, -2) sage: a.real() 3.0 sage: a.real_part() 3.0 sage: a.imag() -2.0 sage: a.imag_part() -2.0 sage: i = ComplexField(100).0 sage: z = 2 + 3*i sage: z.real() 2.0000000000000000000000000000 sage: z.real_part() 2.0000000000000000000000000000 sage: z.imag() 3.0000000000000000000000000000 sage: z.imag_part() 3.0000000000000000000000000000 }}} * Efficient summing using balanced sum (Jason Grout, Mike Hansen) [[http://trac.sagemath.org/sage_trac/ticket/2737|#2737]] --- New function {{{balanced_sum()}}} in the module {{{sage/misc/misc_c.pyx}}} for summing the elements in a list. In some cases, {{{balanced_sum()}}} is more efficient than the built-in Python {{{sum()}}} function, where the efficiency can range from 26x up to 1410x faster than {{{sum()}}}. The following timing statistics were obtained using the machine sage.math: {{{#!python numbers=off sage: R.<x,y> = QQ["x,y"] sage: L = [x^i for i in xrange(1000)] sage: %time sum(L); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s sage: %time balanced_sum(L); CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s Wall time: 0.00 s sage: %timeit sum(L); 100 loops, best of 3: 8.66 ms per loop sage: %timeit balanced_sum(L); 1000 loops, best of 3: 324 µs per loop sage: sage: L = [[i] for i in xrange(10e4)] sage: %time sum(L, []); CPU times: user 84.61 s, sys: 0.00 s, total: 84.61 s Wall time: 84.61 s sage: %time balanced_sum(L, []); CPU times: user 0.06 s, sys: 0.00 s, total: 0.06 s Wall time: 0.06 s }}} |
| Line 22: | Line 81: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/5996|#5996]] | * Wigner `3j`, `6j`, `9j`, Clebsch-Gordan, Racah and Gaunt coefficients (Jens Rasch) [[http://trac.sagemath.org/sage_trac/ticket/5996|#5996]] --- A collection of functions for exactly calculating Wigner [[http://en.wikipedia.org/wiki/3-jm_symbol|`3j`]], [[http://en.wikipedia.org/wiki/6-j_symbol|`6j`]], [[http://en.wikipedia.org/wiki/9-j_symbol|`9j`]], [[http://en.wikipedia.org/wiki/Clebsch-Gordan_coefficients|Clebsch-Gordan]], [[http://en.wikipedia.org/wiki/Racah_W-coefficient|Racah]] as well as Gaunt coefficients. All these functions evaluate to a rational number times the square root of a rational number. These new functions are defined in the module `sage/functions/wigner.py`. Here are some examples on calculating the Wigner `3j`, `6j`, `9j` symbols: {{{#!python numbers=off sage: wigner_3j(2, 6, 4, 0, 0, 0) sqrt(5/143) sage: wigner_3j(0.5, 0.5, 1, 0.5, -0.5, 0) sqrt(1/6) sage: wigner_6j(3,3,3,3,3,3) -1/14 sage: wigner_6j(8,8,8,8,8,8) -12219/965770 sage: wigner_9j(1,1,1, 1,1,1, 1,1,0 ,prec=64) # ==1/18 0.0555555555555555555 sage: wigner_9j(15,15,15, 15,3,15, 15,18,10, prec=1000)*1.0 -0.0000778324615309539 }}} The Clebsch-Gordan, Racah and Gaunt coefficients can be computed as follows: {{{#!python numbers=off sage: simplify(clebsch_gordan(3/2,1/2,2, 3/2,1/2,2)) 1 sage: clebsch_gordan(1.5,0.5,1, 1.5,-0.5,1) 1/2*sqrt(3) sage: racah(3,3,3,3,3,3) -1/14 sage: gaunt(1,0,1,1,0,-1) -1/2/sqrt(pi) sage: gaunt(12,15,5,2,3,-5) 91/124062*sqrt(36890)/sqrt(pi) sage: gaunt(1000,1000,1200,9,3,-12).n(64) 0.00689500421922113448 }}} |
| Line 28: | Line 116: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6519|#6519]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6621|#6621]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/5790|#5790]] |
* Optimize the words library code (Vincent Delecroix, Sébastien Labbé, Franco Saliola) [[http://trac.sagemath.org/sage_trac/ticket/6519|#6519]] --- An enhancement of the words library code in `sage/combinat/words` to improve its efficiency and reorganize the code. The efficiency gain for creating small words can be up to 6x: {{{#!python numbers=off # BEFORE sage: %timeit Word() 10000 loops, best of 3: 46.6 µs per loop sage: %timeit Word("abbabaab") 10000 loops, best of 3: 62 µs per loop sage: %timeit Word([0,1,1,0,1,0,0,1]) 10000 loops, best of 3: 59.4 µs per loop # AFTER sage: %timeit Word() 100000 loops, best of 3: 6.85 µs per loop sage: %timeit Word("abbabaab") 100000 loops, best of 3: 11.8 µs per loop sage: %timeit Word([0,1,1,0,1,0,0,1]) 100000 loops, best of 3: 10.6 µs per loop }}} For the creation of large words, the improvement can be from between 8000x up to 39000x: {{{#!python numbers=off # BEFORE sage: t = words.ThueMorseWord() sage: w = list(t[:1000000]) sage: %timeit Word(w) 10 loops, best of 3: 792 ms per loop sage: u = "".join(map(str, list(t[:1000000]))) sage: %timeit Word(u) 10 loops, best of 3: 777 ms per loop sage: %timeit Words("01")(u) 10 loops, best of 3: 748 ms per loop # AFTER sage: t = words.ThueMorseWord() sage: w = list(t[:1000000]) sage: %timeit Word(w) 10000 loops, best of 3: 20.3 µs per loop sage: u = "".join(map(str, list(t[:1000000]))) sage: %timeit Word(u) 10000 loops, best of 3: 21.9 µs per loop sage: %timeit Words("01")(u) 10000 loops, best of 3: 84.3 µs per loop }}} All of the above timing statistics were obtained using the machine sage.math. Further timing comparisons can be found at the Sage [[http://wiki.sagemath.org/WordDesign|wiki page]]. * Improve the speed of `Permutation.inverse()` (Anders Claesson) [[http://trac.sagemath.org/sage_trac/ticket/6621|#6621]] --- In some cases, the speed gain is up to 11x. The following timing statistics were obtained using the machine sage.math: {{{#!python numbers=off # BEFORE sage: p = Permutation([6, 7, 8, 9, 4, 2, 3, 1, 5]) sage: %timeit p.inverse() 10000 loops, best of 3: 67.1 µs per loop sage: p = Permutation([19, 5, 13, 8, 7, 15, 9, 10, 16, 3, 12, 6, 2, 20, 18, 11, 14, 4, 17, 1]) sage: %timeit p.inverse() 1000 loops, best of 3: 240 µs per loop sage: p = Permutation([14, 17, 1, 24, 16, 34, 19, 9, 20, 18, 36, 5, 22, 2, 27, 40, 37, 15, 3, 35, 10, 25, 21, 8, 13, 26, 12, 32, 23, 38, 11, 4, 6, 39, 31, 28, 29, 7, 30, 33]) sage: %timeit p.inverse() 1000 loops, best of 3: 857 µs per loop # AFTER sage: p = Permutation([6, 7, 8, 9, 4, 2, 3, 1, 5]) sage: %timeit p.inverse() 10000 loops, best of 3: 24.6 µs per loop sage: p = Permutation([19, 5, 13, 8, 7, 15, 9, 10, 16, 3, 12, 6, 2, 20, 18, 11, 14, 4, 17, 1]) sage: %timeit p.inverse() 10000 loops, best of 3: 41.4 µs per loop sage: p = Permutation([14, 17, 1, 24, 16, 34, 19, 9, 20, 18, 36, 5, 22, 2, 27, 40, 37, 15, 3, 35, 10, 25, 21, 8, 13, 26, 12, 32, 23, 38, 11, 4, 6, 39, 31, 28, 29, 7, 30, 33]) sage: %timeit p.inverse() 10000 loops, best of 3: 72.4 µs per loop }}} * Updating some quirks in `sage/combinat/partition.py` (Andrew Mathas) [[http://trac.sagemath.org/sage_trac/ticket/5790|#5790]] --- The functions `r_core()`, `r_quotient()`, `k_core()`, and `partition_sign()` are now deprecated. These are replaced with `core()`, `quotient()`, and `sign()` respectively. The rewrite of the function `Partition()` deprecated the argument `core_and_quotient`. The core and the quotient can be passed as keywords of `Partition()`. {{{#!python numbers=off sage: Partition(core_and_quotient=([2,1], [[2,1],[3],[1,1,1]])) /home/mvngu/.sage/temp/sage.math.washington.edu/9221/_home_mvngu__sage_init_sage_0.py:1: DeprecationWarning: "core_and_quotient=(*)" is deprecated. Use "core=[*], quotient=[*]" instead. # -*- coding: utf-8 -*- [11, 5, 5, 3, 2, 2, 2] sage: Partition(core=[2,1], quotient=[[2,1],[3],[1,1,1]]) [11, 5, 5, 3, 2, 2, 2] sage: Partition([6,3,2,2]).r_quotient(3) /home/mvngu/.sage/temp/sage.math.washington.edu/9221/_home_mvngu__sage_init_sage_0.py:1: DeprecationWarning: r_quotient is deprecated. Please use quotient instead. # -*- coding: utf-8 -*- [[], [], [2, 1]] sage: Partition([6,3,2,2]).quotient(3) [[], [], [2, 1]] sage: partition_sign([5,1,1,1,1,1]) /home/mvngu/.sage/temp/sage.math.washington.edu/9221/_home_mvngu__sage_init_sage_0.py:1: DeprecationWarning: "partition_sign deprecated. Use Partition(pi).sign() instead # -*- coding: utf-8 -*- 1 sage: Partition([5,1,1,1,1,1]).sign() 1 }}} |
| Line 40: | Line 224: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6454|#6454]] == Documentation == * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/4460|#4460]] |
* Improve S-box linear and differences matrices computation (Yann Laigle-Chapuy) [[http://trac.sagemath.org/sage_trac/ticket/6454|#6454]] --- Speed up the functions `difference_distribution_matrix()` and `linear_approximation_matrix()` of the class `SBox` in the module `sage/crypto/mq/sbox.py`. The function `linear_approximation_matrix()` now uses the Walsh transform. The efficiency of `difference_distribution_matrix()` can be up to 277x, while that for `linear_approximation_matrix()` can be up to 132x. The following timing statistics were obtained using the machine sage.math: {{{#!python numbers=off # BEFORE sage: S = mq.SR(1,4,4,8).sbox() sage: %time S.difference_distribution_matrix(); CPU times: user 77.73 s, sys: 0.00 s, total: 77.73 s Wall time: 77.73 s sage: %time S.linear_approximation_matrix(); CPU times: user 132.96 s, sys: 0.00 s, total: 132.96 s Wall time: 132.96 s # AFTER sage: S = mq.SR(1,4,4,8).sbox() sage: %time S.difference_distribution_matrix(); CPU times: user 0.28 s, sys: 0.01 s, total: 0.29 s Wall time: 0.28 s sage: %time S.linear_approximation_matrix(); CPU times: user 1.01 s, sys: 0.00 s, total: 1.01 s Wall time: 1.01 s }}} |
| Line 52: | Line 252: |
| * [[http://trac.sagemath.org/sage_trac/ticket/6381|#6381]] (bug in integral_points when rank is large): The function integral_x_coords_in_interval() for finding all integral points on an elliptic curve defined over the rationals whose x-coordinate lies in an interval is now more efficient when the interval is large. * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6407|#6407]] |
* Allow the method `integral_points()` to handle elliptic curves with large ranks (John Cremona) [[http://trac.sagemath.org/sage_trac/ticket/6381|#6381]] --- A rewrite of the method `integral_x_coords_in_interval()` in the class `EllipticCurve_rational_field` belonging to the module `sage/schemes/elliptic_curves/ell_rational_field.py`. The rewrite allows the method `integral_points()` to compute the integral points of elliptic curves with large ranks. For example, previously the following code would result in an `OverflowError`: {{{#!python numbers=off sage: D = 6611719866 sage: E = EllipticCurve([0, 0, 0, -D^2, 0]) sage: E.integral_points(); }}} * Multiplication-by-n method on elliptic curve formal groups uses the double-and-add algorithm (Hamish Ivey-Law, Tom Boothby) [[http://trac.sagemath.org/sage_trac/ticket/6407|#6407]] --- Previously, the method `EllipticCurveFormalGroup.mult_by_n()` was implemented by applying the group law to itself `n` times. However, when working over a field of characteristic zero, a faster algorithm would be used instead. The linear algorithm is now replaced with the logarithmic double-and-add algorithm, i.e. the additive version of the standard square-and-multiply algorithm. In some cases, the efficiency gain can range from 3% up to 29%. The following timing statistics were obtained using the machine sage.math: {{{#!python numbers=off # BEFORE sage: F = EllipticCurve(GF(101), [1, 1]).formal_group() sage: %time F.mult_by_n(100, 20); CPU times: user 0.98 s, sys: 0.00 s, total: 0.98 s Wall time: 0.98 s sage: F = EllipticCurve("37a").formal_group() sage: %time F.mult_by_n(1000000, 20); CPU times: user 0.38 s, sys: 0.00 s, total: 0.38 s Wall time: 0.38 s sage: %time F.mult_by_n(100000000, 20); CPU times: user 0.55 s, sys: 0.03 s, total: 0.58 s Wall time: 0.58 s # AFTER sage: F = EllipticCurve(GF(101), [1, 1]).formal_group() sage: %time F.mult_by_n(100, 20); CPU times: user 0.96 s, sys: 0.00 s, total: 0.96 s Wall time: 0.95 s sage: F = EllipticCurve("37a").formal_group() sage: %time F.mult_by_n(1000000, 20); CPU times: user 0.44 s, sys: 0.01 s, total: 0.45 s Wall time: 0.45 s sage: %time F.mult_by_n(100000000, 20); CPU times: user 0.40 s, sys: 0.01 s, total: 0.41 s Wall time: 0.41 s }}} |
| Line 62: | Line 296: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6098|#6098]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/5649|#5649]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/5651|#5651]] |
* Plotting 3-D Bezier paths (Emily Kirkman) [[http://trac.sagemath.org/sage_trac/ticket/6098|#6098]] --- New function `bezier3d()` for plotting a 3-dimensional Bezier path. Here are some examples for working with this function: {{{ sage: bezier3d([[(0,0,0), (1,0,0), (0,1,0), (0,1,1)]]).show(zoom=1.2) }}} {{attachment:bezier3d-1.png}} {{{ sage: path = [[(0,0,0),(.5,.1,.2),(.75,3,-1),(1,1,0)],[(.5,1,.2),(1,.5,0)],[(.7,.2,.5)]] sage: bezier3d(path, color='green').show(zoom=1.2) }}} {{attachment:bezier3d-2.png}} * Passing extra options to `show()` (Bill Cauchois) [[http://trac.sagemath.org/sage_trac/ticket/5651|#5651]] --- Extra optional arguments to plotting functions can now be passed on to the function `show()`. This passing of optional arguments is implemented for the following plotting modules: * `sage/plot/arrow.py` * `sage/plot/bar_chart.py` * `sage/plot/bezier_path.py` * `sage/plot/circle.py` * `sage/plot/complex_plot.pyx` * `sage/plot/contour_plot.py` * `sage/plot/density_plot.py` * `sage/plot/disk.py` * `sage/plot/line.py` * `sage/plot/matrix_plot.py` * `sage/plot/plot.py` * `sage/plot/plot_field.py` * `sage/plot/point.py` * `sage/plot/polygon.py` * `sage/plot/scatter_plot.py` * `sage/plot/text.py` Each of the following examples demonstrates equivalent code to obtain a plot: {{{ sage: arrow((-2, 2), (7,1), frame=True) sage: arrow((-2, 2), (7,1)).show(frame=True) }}} {{attachment:arrow.png}} {{{ sage: bar_chart([-2,8,-7,3], rgbcolor=(1,0,0), axes=False) sage: bar_chart([-2,8,-7,3], rgbcolor=(1,0,0)).show(axes=False) }}} {{attachment:bar-chart.png}} {{{ sage: bezier_path([[(0,1),(.5,0),(1,1)]], fontsize=20) sage: bezier_path([[(0,1),(.5,0),(1,1)]]).show(fontsize=20) }}} {{attachment:bezier-path.png}} {{{ sage: complex_plot(lambda z: z, (-3, 3), (-3, 3), figsize=[5,5]) sage: complex_plot(lambda z: z, (-3, 3), (-3, 3)).show(figsize=[5,5]) }}} {{attachment:complex-plot.png}} |
| Line 74: | Line 351: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6355|#6355]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6540|#6540]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6552|#6552]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6578|#6578]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/5793|#5793]] |
* Cliquer as a standard package (Nathann Cohen) [[http://trac.sagemath.org/sage_trac/ticket/6355|#6355]] --- [[http://users.tkk.fi/pat/cliquer.html|Cliquer]] is a set of C routines for finding cliques in an arbitrary weighted graph. It uses an exact branch-and-bound algorithm recently developed by Patric Ostergard and mainly written by Sampo Niskanen. It is published under the GPL license. Here are some examples for working with the new cliquer spkg: {{{#!python numbers=off sage: max_clique(graphs.PetersenGraph()) [7, 9] sage: all_max_clique(graphs.PetersenGraph()) [[2, 7], [7, 9], [6, 8], [6, 9], [0, 4], [4, 9], [5, 7], [0, 5], [5, 8], [3, 4], [2, 3], [3, 8], [1, 6], [0, 1], [1, 2]] sage: clique_number(Graph("DJ{")) 4 sage: clique_number(Graph({0:[1,2,3], 1:[2], 3:[0,1]})) 3 sage: list_composition([1,3,'a'], {'a':'b', 1:2, 2:3, 3:4}) [2, 4, 'b'] }}} * Faster algorithm to compute maximum cliques (Nathann Cohen) [[http://trac.sagemath.org/sage_trac/ticket/5793|#5793]] --- With the inclusion of cliquer as a standard spkg, the following functions can now use the cliquer algorithm: * `Graph.max_clique()` --- Returns the vertex set of a maximum complete subgraph. * `Graph.cliques_maximum()` --- Returns the list of all maximum cliques, with each clique represented by a list of vertices. A clique is an induced complete subgraph and a maximal clique is one of maximal order. * `Graph.clique_number()` --- Returns the size of the largest clique of the graph. * `Graph.cliques_vertex_clique_number()` --- Returns a list of sizes of the largest maximal cliques containing each vertex. This returns a single value if there is only one input vertex. * `Graph.independent_set()` --- Returns a maximal independent set, which is a set of vertices which induces an empty subgraph. These functions already exist in Sage. Cliquer does not bring to Sage any new feature, but a huge efficiency improvement in computing clique numbers. The NetworkX 0.36 algorithm is very slow in its computation of these functions, even though it remains faster than cliquer for the computation of `Graph.cliques_vertex_clique_number()`. The algorithms in the cliquer spkg scale very well as the number of vertices in a graph increases. Here is a comparison between the implementation of NetworkX 0.36 and cliquer on computing the clique number of a graph. Timing statistics were obtained using the machine sage.math: {{{#!python numbers=off sage: g = graphs.RandomGNP(100, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 0.64 s, sys: 0.01 s, total: 0.65 s Wall time: 0.65 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 0.02 s, sys: 0.00 s, total: 0.02 s Wall time: 0.02 s sage: g = graphs.RandomGNP(200, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 9.68 s, sys: 0.01 s, total: 9.69 s Wall time: 9.68 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 0.09 s, sys: 0.00 s, total: 0.09 s Wall time: 0.09 s sage: g = graphs.RandomGNP(300, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 69.98 s, sys: 0.10 s, total: 70.08 s Wall time: 70.09 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 0.23 s, sys: 0.00 s, total: 0.23 s Wall time: 0.23 s sage: g = graphs.RandomGNP(400, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 299.32 s, sys: 0.29 s, total: 299.61 s Wall time: 299.64 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 0.54 s, sys: 0.00 s, total: 0.54 s Wall time: 0.53 s sage: g = graphs.RandomGNP(500, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 1178.85 s, sys: 1.30 s, total: 1180.15 s Wall time: 1180.16 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 1.09 s, sys: 0.00 s, total: 1.09 s Wall time: 1.09 s }}} * Support the syntax `g.add_edge((u,v), label=l)` for C graphs (Robert Miller) [[http://trac.sagemath.org/sage_trac/ticket/6540|#6540]] --- The following syntax is supported. However, note that the `label` keyword must be used: {{{#!python numbers=off sage: G = Graph() sage: G.add_edge((1,2), label="my label") sage: G.edges() [(1, 2, 'my label')] sage: G = Graph() sage: G.add_edge((1,2), "label") sage: G.edges() [((1, 2), 'label', None)] }}} * Fast subgraphs by building the graph instead of deleting things (Jason Grout) [[http://trac.sagemath.org/sage_trac/ticket/6578|#6578]] --- Subgraphs can now be constructed by building a new graph from a number of vertices and edges. This is in contrast to the previous default algorithm where subgraphs were constructed by deleting edges and vertices. In some cases, the efficiency gain of the new subgraph construction implementation can be up to 17x. The following timing statistics were obtained using the machine sage.math: {{{#!python numbers=off # BEFORE sage: g = graphs.PathGraph(Integer(10e4)) sage: %time g.subgraph(range(20)); CPU times: user 1.89 s, sys: 0.03 s, total: 1.92 s Wall time: 1.92 s sage: g = graphs.PathGraph(Integer(10e4) * 5) sage: %time g.subgraph(range(20)); CPU times: user 14.92 s, sys: 0.05 s, total: 14.97 s Wall time: 14.97 s sage: g = graphs.PathGraph(Integer(10e5)) sage: %time g.subgraph(range(20)); CPU times: user 47.77 s, sys: 0.29 s, total: 48.06 s Wall time: 48.06 s # AFTER sage: g = graphs.PathGraph(Integer(10e4)) sage: %time g.subgraph(range(20)); CPU times: user 0.27 s, sys: 0.01 s, total: 0.28 s Wall time: 0.28 s sage: g = graphs.PathGraph(Integer(10e4) * 5) sage: %time g.subgraph(range(20)); CPU times: user 1.34 s, sys: 0.03 s, total: 1.37 s Wall time: 1.37 s sage: g = graphs.PathGraph(Integer(10e5)) sage: %time g.subgraph(range(20)); CPU times: user 2.66 s, sys: 0.04 s, total: 2.70 s Wall time: 2.70 s }}} |
| Line 92: | Line 483: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6395|#6395]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6591|#6591]] |
* Magma interface: make `magma_colon_equals()` mode work in both command line and notebook (William Stein) [[http://trac.sagemath.org/sage_trac/ticket/6395|#6395]] --- Exposes the `magma_colon_equals()` option in the notebook. For example, one can now do the following in the notebook: {{{#!python numbers=off sage: magma._preparse_colon_equals = False sage: magma._preparse('a = 5') 'a = 5' sage: magma._preparse_colon_equals = True sage: magma._preparse('a = 5') 'a := 5' sage: magma._preparse('a = 5; b := 7; c =a+b;') 'a := 5; b := 7; c :=a+b;' }}} * Viewing a Sage object with `view(object, viewer='pdf')` (Nicolas M. Thiéry) [[http://trac.sagemath.org/sage_trac/ticket/6591|#6591]] --- Typical uses include: * you prefer your PDF browser to your DVI browser * you want to view LaTeX snippets which are not displayed well in DVI viewers or jsMath, e.g. images produced by tikzpicture. |
| Line 101: | Line 504: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/5081|#5081]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6553|#6553]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6554|#6554]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6301|#6301]] |
* Make NumPy play nice with Sage types (Robert Bradshaw) [[http://trac.sagemath.org/sage_trac/ticket/5081|#5081]] --- This improves data conversion between NumPy and Sage. For example, one can now do this: {{{#!python numbers=off sage: from scipy import stats sage: stats.uniform(0,15).ppf([0.5,0.7]) array([ 7.5, 10.5]) }}} And this: {{{#!python numbers=off sage: from scipy import * sage: from pylab import * sage: sample_rate = 1000.0 sage: t = r_[0:0.6:1/sample_rate] sage: N = len(t) sage: s = [sin(2*pi*50*elem) + sin(2*pi*70*elem + (pi/4)) for elem in t] sage: S = fft(s) sage: f = sample_rate*r_[0:(N/2)] / N sage: n = len(f) sage: line(zip(f, abs(S[0:n]) / N)) }}} {{attachment:fft.png}} * Fast slicing of sparse matrices (Jason Grout) [[http://trac.sagemath.org/sage_trac/ticket/6553|#6553]] --- The efficiency gain for slicing sparse matrices can range from 10x up to 147x. The following timing statistics were obtained using the machine sage.math: {{{#!python numbers=off # BEFORE sage: A = random_matrix(ZZ, 100000, density=0.00005, sparse=True) sage: %time A[50000:,:50000]; CPU times: user 298.84 s, sys: 0.05 s, total: 298.89 s Wall time: 298.95 s sage: A = random_matrix(ZZ, 10000, density=0.00005, sparse=True) sage: %time A[5000:,:5000]; CPU times: user 2.50 s, sys: 0.00 s, total: 2.50 s Wall time: 2.50 s # AFTER sage: A = random_matrix(ZZ, 100000, density=0.00005, sparse=True) sage: %time A[50000:,:50000]; CPU times: user 1.91 s, sys: 0.09 s, total: 2.00 s Wall time: 2.02 s sage: A = random_matrix(ZZ, 10000, density=0.00005, sparse=True) sage: %time A[5000:,:5000]; CPU times: user 0.23 s, sys: 0.00 s, total: 0.23 s Wall time: 0.24 s }}} * Plotting sparse matrices efficiently (Jason Grout) [[http://trac.sagemath.org/sage_trac/ticket/6554|#6554]] --- Previously, plotting a sparse matrix would convert the matrix to a dense matrix, resulting in the whole process taking increasingly longer time as the dimensions of the matrix increase. Where a matrix is sparse, the matrix plotting function now uses SciPy's sparse matrix functionalities, which can handle large matrices. In some cases, the performance improvement can range from 380x up to 98000x. The following timing statistics were obtained using the machine mod.math: {{{#!python numbers=off # BEFORE sage: A = random_matrix(ZZ, 5000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 60.25 s, sys: 0.69 s, total: 60.94 s Wall time: 60.94 s sage: A = random_matrix(ZZ, 10000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 241.31 s, sys: 3.03 s, total: 244.34 s Wall time: 244.35 s sage: A = random_matrix(ZZ, 15000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 544.02 s, sys: 6.85 s, total: 550.87 s Wall time: 550.86 s sage: A = random_matrix(ZZ, 20000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 972.85 s, sys: 13.36 s, total: 986.21 s Wall time: 986.21 s # AFTER sage: A = random_matrix(ZZ, 5000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.05 s, sys: 0.04 s, total: 0.09 s Wall time: 0.16 s sage: A = random_matrix(ZZ, 10000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s sage: A = random_matrix(ZZ, 15000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s sage: A = random_matrix(ZZ, 20000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s }}} In Sage 4.1, the following would quickly consume gigabytes of RAM on a system and may result in a `MemoryError`: {{{#!python numbers=off sage: A = random_matrix(ZZ, 100000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.63 s, sys: 0.01 s, total: 0.64 s Wall time: 0.63 s sage: A = random_matrix(ZZ, 1000000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 1933.41 s, sys: 2.97 s, total: 1936.38 s Wall time: 1937.31 s }}} * Elementwise (Hadamard) product of matrices (Rob Beezer) [[http://trac.sagemath.org/sage_trac/ticket/6301|#6301]] --- Given matrices `A` and `B` of the same size, `C = A.elementwise_product(B)` creates the new matrix C, of the same size, with entries given by `C[i,j] = A[i,j] * B[i,j]`. The multiplication occurs in a ring defined by Sage's coercion model, as appropriate for the base rings of `A` and `B` (or an error is raised if there is no sensible common ring). In particular, if `A` and `B` are defined over a noncommutative ring, the operation is also noncommutative. The implementation is different for dense matrices versus sparse matrices, but there are probably further optimizations available for specific rings. This operation is often called the Hadamard product. Here is an example on using elementwise matrix product: {{{#!python numbers=off sage: G = matrix(GF(3), 2, [0,1,2,2]) sage: H = matrix(ZZ, 2, [1,2,3,4]) sage: J = G.elementwise_product(H); J [0 2] [0 2] sage: J.parent() Full MatrixSpace of 2 by 2 dense matrices over Finite Field of size 3 }}} |
| Line 116: | Line 622: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6606|#6606]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6071|#6071]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6534|#6534]] |
* Efficient implementation of index for `Gamma(N)` (Simon Morris) [[http://trac.sagemath.org/sage_trac/ticket/6606|#6606]] --- The new implementation provides massive speed-up for the function `Gamma(N)`. The following statistics were obtained using the machine mod.math: {{{#!python numbers=off # BEFORE sage: %time [Gamma(n).index() for n in [1..19]]; CPU times: user 14369.53 s, sys: 75.18 s, total: 14444.71 s Wall time: 14445.22 s sage: %time Gamma(32041).index(); # hangs for hours # AFTER sage: %time [Gamma(n).index() for n in [1..19]]; CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s Wall time: 0.00 s sage: timeit("[Gamma(n).index() for n in [1..19]]") 125 loops, best of 3: 2.27 ms per loop sage: %time Gamma(32041).index(); CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s sage: %timeit Gamma(32041).index() 10000 loops, best of 3: 110 µs per loop }}} * Weight 1 Eisenstein series (David Loeffler) [[http://trac.sagemath.org/sage_trac/ticket/6071|#6071]] --- Add support for computing weight 1 Eisenstein series. Here's an example: {{{#!python numbers=off sage: M = ModularForms(DirichletGroup(13).0, 1) sage: M.eisenstein_series() [ -1/13*zeta12^3 + 6/13*zeta12^2 + 4/13*zeta12 + 2/13 + q + (zeta12 + 1)*q^2 + zeta12^2*q^3 + (zeta12^2 + zeta12 + 1)*q^4 + (-zeta12^3 + 1)*q^5 + O(q^6) ] }}} * Efficient calculation of Jacobi sums (David Loeffler) [[http://trac.sagemath.org/sage_trac/ticket/6534|#6534]] --- For small primes, calculating Jacobi sums using the definition directly can result in an efficiency improvement of up to 624x. The following timing statistics were obtained using the machine mod.math: {{{#!python numbers=off # BEFORE sage: chi = DirichletGroup(67).0 sage: psi = chi**3 sage: time chi.jacobi_sum(psi); CPU times: user 6.24 s, sys: 0.00 s, total: 6.24 s Wall time: 6.24 s # AFTER sage: chi = DirichletGroup(67).0 sage: psi = chi**3 sage: time chi.jacobi_sum(psi); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s }}} There is also support for computing a Jacobi sum with values in a finite field: {{{#!python numbers=off sage: g = DirichletGroup(17, GF(9,'a')).0 sage: g.jacobi_sum(g**2) 2*a }}} |
| Line 128: | Line 685: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/5653|#5653]] | * Display docstrings in the notebook using HTML and jsMath (Tom Boothby, Evan Fosmark, John Palmieri, Mitesh Patel) [[http://trac.sagemath.org/sage_trac/ticket/5653|#5653]] --- When viewing docstrings using the notebook, these are presented using HTML and jsMath. For example, if one does any of the following {{{ identity_matrix([TAB] identity_matrix?[SHIFT-RETURN] identity_matrix?[TAB] }}} then the docstring for the function `identity_matrix()` would be presented as in this figure: {{attachment:notebook-docstring.png}} |
| Line 134: | Line 699: |
| * [[http://trac.sagemath.org/sage_trac/ticket/6457|#6457]] (Intersection of ideals in a number field) Intersection of ideals in number fields is now implemented. * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6045|#6045]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6396|#6396]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6458|#6458]] |
* Intersection of ideals in a number field (David Loeffler) [[http://trac.sagemath.org/sage_trac/ticket/6457|#6457]] --- New function `intersection()` of the class `NumberFieldIdeal` in the module `sage/rings/number_field/number_field_ideal.py` for computing the ideals in a number field. Here are some examples on using `intersection()`: {{{#!python numbers=off sage: K.<a> = QuadraticField(-11) sage: p = K.ideal((a + 1)/2); q = K.ideal((a + 3)/2) sage: p.intersection(q) == q.intersection(p) == K.ideal(a-2) True sage: # An example with non-principal ideals sage: L.<a> = NumberField(x^3 - 7) sage: p = L.ideal(a^2 + a + 1, 2) sage: q = L.ideal(a+1) sage: p.intersection(q) == L.ideal(8, 2*a + 2) True sage: # A relative example sage: L.<a,b> = NumberField([x^2 + 11, x^2 - 5]) sage: A = L.ideal([15, (-3/2*b + 7/2)*a - 8]) sage: B = L.ideal([6, (-1/2*b + 1)*a - b - 5/2]) sage: A.intersection(B) == L.ideal(-1/2*a - 3/2*b - 1) True }}} * Computing Heegner points (Robert Bradshaw) [[http://trac.sagemath.org/sage_trac/ticket/6045|#6045]] --- Adds a Heegner point method to elliptic curves over `QQ`. The new method is `heegner_point()` which can be found in the class `EllipticCurve_rational_field` of the module `sage/schemes/elliptic_curves/ell_rational_field.py`. Here are some examples on using `heegner_point()`: {{{#!python numbers=off sage: E = EllipticCurve('37a') sage: E.heegner_discriminants_list(10) [-7, -11, -40, -47, -67, -71, -83, -84, -95, -104] sage: E.heegner_point(-7) (0 : 0 : 1) sage: P = E.heegner_point(-40); P (a : -a + 1 : 1) sage: P = E.heegner_point(-47); P (a : -a^4 - a : 1) sage: P[0].parent() Number Field in a with defining polynomial x^5 - x^4 + x^3 + x^2 - 2*x + 1 sage: # Working out the details manually sage: P = E.heegner_point(-47, prec=200) sage: f = algdep(P[0], 5); f x^5 - x^4 + x^3 + x^2 - 2*x + 1 sage: f.discriminant().factor() 47^2 }}} * Support `primes_of_degree_one()` for relative extensions (David Loeffler) [[http://trac.sagemath.org/sage_trac/ticket/6396|#6396]] --- For example, one can now do this: {{{#!python numbers=off sage: N.<a,b> = NumberField([x^2 + 1, x^2 - 5]) sage: ids = N.primes_of_degree_one_list(10); ids [Fractional ideal (2*a + 1/2*b - 1/2), Fractional ideal ((-1/2*b - 1/2)*a - b), Fractional ideal (b*a + 1/2*b + 3/2), Fractional ideal ((-1/2*b - 3/2)*a + b - 1), Fractional ideal ((-b + 1)*a + b), Fractional ideal (3*a + 1/2*b - 1/2), Fractional ideal ((1/2*b - 1/2)*a + 3/2*b - 1/2), Fractional ideal ((-1/2*b - 5/2)*a - b + 1), Fractional ideal (2*a - 3/2*b - 1/2), Fractional ideal (3*a + 1/2*b + 5/2)] sage: [x.absolute_norm() for x in ids] [29, 41, 61, 89, 101, 109, 149, 181, 229, 241] sage: ids[9] == N.ideal(3*a + 1/2*b + 5/2) True }}} * Inverse modulo an ideal in a relative number field (David Loeffler) [[http://trac.sagemath.org/sage_trac/ticket/6458|#6458]] --- Support for computing the inverse modulo an ideal in the ring of integers of a relative field. Here's an example: {{{#!python numbers=off sage: K.<a,b> = NumberField([x^2 + 1, x^2 - 3]) sage: I = K.ideal(17*b - 3*a) sage: x = I.integral_basis(); x [438, -b*a + 309, 219*a - 219*b, 156*a - 154*b] sage: V, _, phi = K.absolute_vector_space() sage: V.span([phi(u) for u in x], ZZ) == I.free_module() True }}} |
| Line 150: | Line 779: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6506|#6506]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/4571|#4571]] |
* Further NumPy type conversions (Robert Bradshaw, Jason Grout) [[http://trac.sagemath.org/sage_trac/ticket/6506|#6506]] --- Support improved handling of `ZZ`, `QQ`, and `CC` between NumPy and Sage. Here are some examples: {{{#!python numbers=off sage: import numpy sage: numpy.array([1.0, 2.5j]) array([ 1.+0.j , 0.+2.5j]) sage: numpy.array([1.0, 2.5j]).dtype dtype('complex128') sage: numpy.array([1.000000000000000000000000000000000000j]).dtype dtype('object') sage: numpy.array([1, 2, 3]) array([1, 2, 3]) sage: numpy.array([1, 2, 3]).dtype dtype('int64') sage: numpy.array(2**40).dtype dtype('int64') sage: numpy.array(2**400).dtype dtype('object') sage: numpy.array([1,2,3,0.1]).dtype dtype('float64') sage: numpy.array(QQ(2**40)).dtype dtype('int64') sage: numpy.array(QQ(2**400)).dtype dtype('object') sage: numpy.array([1, 1/2, 3/4]) array([ 1. , 0.5 , 0.75]) }}} |
| Line 159: | Line 810: |
| * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6558|#6558]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6380|#6380]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6443|#6443]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6445|#6445]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6451|#6451]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6453|#6453]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6528|#6528]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6143|#6143]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6438|#6438]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6493|#6493]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6563|#6563]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6602|#6602]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6302|#6302]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6491|#6491]] == Symbolics == * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6195|#6195]] * FIXME: summarize [[http://trac.sagemath.org/sage_trac/ticket/6243|#6243]] |
* Update the [[http://math-atlas.sourceforge.net|ATLAS]] spkg to version 3.8.3.p7 (David Kirkby, Minh Van Nguyen) [[http://trac.sagemath.org/sage_trac/ticket/6558|#6558]] [[http://trac.sagemath.org/sage_trac/ticket/6738|#6738]] --- This adds support for building ATLAS under Solaris on a Sun SPARC processor. * Update the [[http://www.shoup.net/ntl|NTL]] spkg to version 5.4.2.p9 (David Kirkby) [[http://trac.sagemath.org/sage_trac/ticket/6380|#6380]] --- This adds support for building NTL under Solaris on a Sun SPARC processor. * Update the [[http://cims.nyu.edu/~harvey/zn_poly|zn_poly]] spkg to version 0.9.p1 (David Kirkby) [[http://trac.sagemath.org/sage_trac/ticket/6443|#6443]] --- This adds support for building zn_poly under Solaris on a Sun SPARC processor. * Update the [[http://pari.math.u-bordeaux.fr|Pari/GP]] spkg to version 2.3.3.p1 (David Kirkby) [[http://trac.sagemath.org/sage_trac/ticket/6445|#6445]] --- This adds support for building Pari under Solaris on a Sun SPARC processor. * Update the [[http://www.flintlib.org|FLINT]] spkg to version 1.3.0.p2 (David Kirkby) [[http://trac.sagemath.org/sage_trac/ticket/6451|#6451]] --- This fixes a Solaris specific bug in FLINT. * Update the [[http://www.mpfr.org|MPFR]] spkg to version 2.4.1.p0 (Paul Zimmermann, David Kirkby) [[http://trac.sagemath.org/sage_trac/ticket/6453|#6453]] --- This fixes a number of test failures under Solaris on a Sun SPARC processor. * Update the [[http://polybori.sourceforge.net|PolyBoRi]] spkg to version 0.5rc.p9 (David Kirkby) [[http://trac.sagemath.org/sage_trac/ticket/6528|#6528]] --- This fixes a Solaris specific bug in compiling PolyBoRi with the Sun compiler. * Upgrade [[http://tinymce.moxiecode.com|tinyMCE]] to version 3.2.4.1 upstream release (Jason Grout) [[http://trac.sagemath.org/sage_trac/ticket/6143|#6143]] --- This version of tinyMCE has many fixes for Safari and a greatly improved paste-from-word functionality. * Upgrade [[http://www.cython.org|Cython]] to version 0.11.2.1 latest upstream release (Robert Bradshaw) [[http://trac.sagemath.org/sage_trac/ticket/6438|#6438]]. * Update the [[http://numpy.scipy.org|NumPy]] spkg to version 1.3.0.p1 (William Stein) [[http://trac.sagemath.org/sage_trac/ticket/6493|#6493]] --- This fixes a bug in compiling NumPy under 64-bit OS X. * Upgrade [[http://www.singular.uni-kl.de|Singular]] to version singular-3-1-0-2-20090620.p0 (David Kirkby) [[http://trac.sagemath.org/sage_trac/ticket/6563|#6563]] --- This fixes a Solaris specific bug when compiling Singular under Solaris on a Sun SPARC processor. * New optional spkg [[http://www.gnu.org/software/glpk|GLPK]] version 4.38 (Nathann Cohen) [[http://trac.sagemath.org/sage_trac/ticket/6602|#6602]] --- GLPK is a linear program solver that can also solve mixed integer programs. * New optional spkg [[http://www.openopt.org|OpenOpt]] version 0.24 (William Stein) [[http://trac.sagemath.org/sage_trac/ticket/6302|#6302]] --- OpenOpt is a numerical optimization with various solvers. * New optional package [[http://sage.math.washington.edu/home/SimonKing/Cohomology/|p_group_cohomology]] version 1.0.2 (Simon A. King, David J. Green) [[http://trac.sagemath.org/sage_trac/ticket/6491|#6491]] --- The package p_group_cohomology can compute the cohomology ring of a group with coefficients in a finite field of order p. Its features include: * Compute the cohomology ring with coefficients in GF(p) for any finite p-group, in terms of a minimal generating set and a minimal set of algebraic relations. We use Benson's criterion to prove the completeness of the ring structure. * Compute depth, dimension, Poincare series and a-invariants of the cohomology rings. * Compute the nil radical * Construct induced homomorphisms. * The package includes a list of cohomology rings for all groups of order 64. * With the package, the cohomology for all groups of order 128 and for the Sylow 2-subgroup of the third Conway group (order 1024) was computed for the first time. The result of these and many other computations (e.g., all but 6 groups of order 243) is accessible in a repository on sage.math. Here some examples: * Data that are included with the package: {{{#!python numbers=off sage: from pGroupCohomology import CohomologyRing sage: H = CohomologyRing(64,132) # this is included in the package, hence, the ring structure is already there sage: print H Cohomology ring of Small Group number 132 of order 64 with coefficients in GF(2) Computation complete Minimal list of generators: [a_2_4, a 2-Cochain in H^*(SmallGroup(64,132); GF(2)), c_2_5, a 2-Cochain in H^*(SmallGroup(64,132); GF(2)), c_4_12, a 4-Cochain in H^*(SmallGroup(64,132); GF(2)), a_1_0, a 1-Cochain in H^*(SmallGroup(64,132); GF(2)), a_1_1, a 1-Cochain in H^*(SmallGroup(64,132); GF(2)), b_1_2, a 1-Cochain in H^*(SmallGroup(64,132); GF(2))] Minimal list of algebraic relations: [a_1_0*a_1_1, a_1_0*b_1_2, a_1_1^3+a_1_0^3, a_2_4*a_1_0, a_1_1^2*b_1_2^2+a_2_4*a_1_1*b_1_2+a_2_4^2+c_2_5*a_1_1^2] sage: H.depth() 2 sage: H.a_invariants() [-Infinity, -Infinity, -3, -3] sage: H.poincare_series() (-t^2 - t - 1)/(t^6 - 2*t^5 + t^4 - t^2 + 2*t - 1) sage: H.nil_radical() a_1_0, a_1_1, a_2_4 }}} * Data from the repository on sage.math: {{{#!python numbers=off sage: H = CohomologyRing(128,562) # if there is internet connection, the ring data are downloaded behind the scenes sage: len(H.gens()) 35 sage: len(H.rels()) 486 sage: H.depth() 1 sage: H.a_invariants() [-Infinity, -4, -3, -3] sage: H.poincare_series() (t^14 - 2*t^13 + 2*t^12 - t^11 - t^10 + t^9 - 2*t^8 + 2*t^7 - 2*t^6 + 2*t^5 - 2*t^4 + t^3 - t^2 - 1)/(t^17 - 3*t^16 + 4*t^15 - 4*t^14 + 4*t^13 - 4*t^12 + 4*t^11 - 4*t^10 + 4*t^9 - 4*t^8 + 4*t^7 - 4*t^6 + 4*t^5 - 4*t^4 + 4*t^3 - 4*t^2 + 3*t - 1) }}} * Some computation from scratch, involving different ring presentations and induced maps: {{{#!python numbers=off sage: tmp_root = tmp_filename() sage: CohomologyRing.set_user_db(tmp_root) sage: H0 = CohomologyRing.user_db(8,3,websource=False) sage: print H0 Cohomology ring of Dihedral group of order 8 with coefficients in GF(2) Computed up to degree 0 Minimal list of generators: [] Minimal list of algebraic relations: [] sage: H0.make() sage: print H0 Cohomology ring of Dihedral group of order 8 with coefficients in GF(2) Computation complete Minimal list of generators: [c_2_2, a 2-Cochain in H^*(D8; GF(2)), b_1_0, a 1-Cochain in H^*(D8; GF(2)), b_1_1, a 1-Cochain in H^*(D8; GF(2))] Minimal list of algebraic relations: [b_1_0*b_1_1] sage: G = gap('DihedralGroup(8)') sage: H1 = CohomologyRing.user_db(G,GroupName='GapD8',websource=False) sage: H1.make() sage: print H1 # the ring presentation is different ... Cohomology ring of GapD8 with coefficients in GF(2) Computation complete Minimal list of generators: [c_2_2, a 2-Cochain in H^*(GapD8; GF(2)), b_1_0, a 1-Cochain in H^*(GapD8; GF(2)), b_1_1, a 1-Cochain in H^*(GapD8; GF(2))] Minimal list of algebraic relations: [b_1_1^2+b_1_0*b_1_1] sage: phi = G.IsomorphismGroups(H0.group()) sage: phi_star = H0.hom(phi,H1) sage: phi_star_inv = phi_star^(-1) # ... but the rings are isomorphic sage: [X==phi_star_inv(phi_star(X)) for X in H0.gens()] [True, True, True, True] sage: [X==phi_star(phi_star_inv(X)) for X in H1.gens()] [True, True, True, True] }}} * An example with an odd prime: {{{#!python numbers=off sage: H = CohomologyRing(81,8) # this needs to be computed from scratch sage: H.make() sage: H.gens() [1, a_2_1, a 2-Cochain in H^*(SmallGroup(81,8); GF(3)), a_2_2, a 2-Cochain in H^*(SmallGroup(81,8); GF(3)), b_2_0, a 2-Cochain in H^*(SmallGroup(81,8); GF(3)), a_4_1, a 4-Cochain in H^*(SmallGroup(81,8); GF(3)), b_4_2, a 4-Cochain in H^*(SmallGroup(81,8); GF(3)), b_6_3, a 6-Cochain in H^*(SmallGroup(81,8); GF(3)), c_6_4, a 6-Cochain in H^*(SmallGroup(81,8); GF(3)), a_1_0, a 1-Cochain in H^*(SmallGroup(81,8); GF(3)), a_1_1, a 1-Cochain in H^*(SmallGroup(81,8); GF(3)), a_3_2, a 3-Cochain in H^*(SmallGroup(81,8); GF(3)), a_5_2, a 5-Cochain in H^*(SmallGroup(81,8); GF(3)), a_5_3, a 5-Cochain in H^*(SmallGroup(81,8); GF(3)), a_7_5, a 7-Cochain in H^*(SmallGroup(81,8); GF(3))] sage: len(H.rels()) 59 sage: H.depth() 1 sage: H.a_invariants() [-Infinity, -3, -2] sage: H.poincare_series() (t^4 - t^3 + t^2 + 1)/(t^6 - 2*t^5 + 2*t^4 - 2*t^3 + 2*t^2 - 2*t + 1) sage: H.nil_radical() a_1_0, a_1_1, a_2_1, a_2_2, a_3_2, a_4_1, a_5_2, a_5_3, b_2_0*b_4_2, a_7_5, b_2_0*b_6_3, b_6_3^2+b_4_2^3 }}} |
Sage 4.1.1 Release Tour
Sage 4.1.1 was released on August 14th, 2009. For the official, comprehensive release note, please refer to sage-4.1.1.txt. A nicely formatted version of this release tour can be found here. The following points are some of the foci of this release:
Improved data conversion between NumPy and Sage
- Solaris support, Solaris specific bug fixes for NTL, zn_poly, Pari/GP, FLINT, MPFR, PolyBoRI, ATLAS
- Upgrade/updates for about 8 standard packages
- Three new optional packages: openopt, GLPK, p_group_cohomology
Algebra
Adds method __nonzero__() to abelian groups (Taylor Sutton) #6510 --- New method __nonzero__() for the class AbelianGroup_class in sage/groups/abelian_gps/abelian_group.py. This method returns True if the abelian group in question is non-trivial:
sage: E = EllipticCurve([0, 82]) sage: T = E.torsion_subgroup() sage: bool(T) False sage: T.__nonzero__() False
Basic Arithmetic
Implement real_part() and imag_part() for CDF and CC (Alex Ghitza) #6159 --- The name real_part is now an alias to the method real(); similarly, imag_part is now an alias to the method imag().
sage: a = CDF(3, -2) sage: a.real() 3.0 sage: a.real_part() 3.0 sage: a.imag() -2.0 sage: a.imag_part() -2.0 sage: i = ComplexField(100).0 sage: z = 2 + 3*i sage: z.real() 2.0000000000000000000000000000 sage: z.real_part() 2.0000000000000000000000000000 sage: z.imag() 3.0000000000000000000000000000 sage: z.imag_part() 3.0000000000000000000000000000
Efficient summing using balanced sum (Jason Grout, Mike Hansen) #2737 --- New function balanced_sum() in the module sage/misc/misc_c.pyx for summing the elements in a list. In some cases, balanced_sum() is more efficient than the built-in Python sum() function, where the efficiency can range from 26x up to 1410x faster than sum(). The following timing statistics were obtained using the machine sage.math:
sage: R.<x,y> = QQ["x,y"] sage: L = [x^i for i in xrange(1000)] sage: %time sum(L); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s sage: %time balanced_sum(L); CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s Wall time: 0.00 s sage: %timeit sum(L); 100 loops, best of 3: 8.66 ms per loop sage: %timeit balanced_sum(L); 1000 loops, best of 3: 324 µs per loop sage: sage: L = [[i] for i in xrange(10e4)] sage: %time sum(L, []); CPU times: user 84.61 s, sys: 0.00 s, total: 84.61 s Wall time: 84.61 s sage: %time balanced_sum(L, []); CPU times: user 0.06 s, sys: 0.00 s, total: 0.06 s Wall time: 0.06 s
Calculus
Wigner 3j, 6j, 9j, Clebsch-Gordan, Racah and Gaunt coefficients (Jens Rasch) #5996 --- A collection of functions for exactly calculating Wigner `3j`, `6j`, `9j`, Clebsch-Gordan, Racah as well as Gaunt coefficients. All these functions evaluate to a rational number times the square root of a rational number. These new functions are defined in the module sage/functions/wigner.py. Here are some examples on calculating the Wigner 3j, 6j, 9j symbols:
The Clebsch-Gordan, Racah and Gaunt coefficients can be computed as follows:sage: wigner_3j(2, 6, 4, 0, 0, 0) sqrt(5/143) sage: wigner_3j(0.5, 0.5, 1, 0.5, -0.5, 0) sqrt(1/6) sage: wigner_6j(3,3,3,3,3,3) -1/14 sage: wigner_6j(8,8,8,8,8,8) -12219/965770 sage: wigner_9j(1,1,1, 1,1,1, 1,1,0 ,prec=64) # ==1/18 0.0555555555555555555 sage: wigner_9j(15,15,15, 15,3,15, 15,18,10, prec=1000)*1.0 -0.0000778324615309539
sage: simplify(clebsch_gordan(3/2,1/2,2, 3/2,1/2,2)) 1 sage: clebsch_gordan(1.5,0.5,1, 1.5,-0.5,1) 1/2*sqrt(3) sage: racah(3,3,3,3,3,3) -1/14 sage: gaunt(1,0,1,1,0,-1) -1/2/sqrt(pi) sage: gaunt(12,15,5,2,3,-5) 91/124062*sqrt(36890)/sqrt(pi) sage: gaunt(1000,1000,1200,9,3,-12).n(64) 0.00689500421922113448
Combinatorics
Optimize the words library code (Vincent Delecroix, Sébastien Labbé, Franco Saliola) #6519 --- An enhancement of the words library code in sage/combinat/words to improve its efficiency and reorganize the code. The efficiency gain for creating small words can be up to 6x:
For the creation of large words, the improvement can be from between 8000x up to 39000x:# BEFORE sage: %timeit Word() 10000 loops, best of 3: 46.6 µs per loop sage: %timeit Word("abbabaab") 10000 loops, best of 3: 62 µs per loop sage: %timeit Word([0,1,1,0,1,0,0,1]) 10000 loops, best of 3: 59.4 µs per loop # AFTER sage: %timeit Word() 100000 loops, best of 3: 6.85 µs per loop sage: %timeit Word("abbabaab") 100000 loops, best of 3: 11.8 µs per loop sage: %timeit Word([0,1,1,0,1,0,0,1]) 100000 loops, best of 3: 10.6 µs per loop
# BEFORE sage: t = words.ThueMorseWord() sage: w = list(t[:1000000]) sage: %timeit Word(w) 10 loops, best of 3: 792 ms per loop sage: u = "".join(map(str, list(t[:1000000]))) sage: %timeit Word(u) 10 loops, best of 3: 777 ms per loop sage: %timeit Words("01")(u) 10 loops, best of 3: 748 ms per loop # AFTER sage: t = words.ThueMorseWord() sage: w = list(t[:1000000]) sage: %timeit Word(w) 10000 loops, best of 3: 20.3 µs per loop sage: u = "".join(map(str, list(t[:1000000]))) sage: %timeit Word(u) 10000 loops, best of 3: 21.9 µs per loop sage: %timeit Words("01")(u) 10000 loops, best of 3: 84.3 µs per loop
All of the above timing statistics were obtained using the machine sage.math. Further timing comparisons can be found at the Sage wiki page.
Improve the speed of Permutation.inverse() (Anders Claesson) #6621 --- In some cases, the speed gain is up to 11x. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: p = Permutation([6, 7, 8, 9, 4, 2, 3, 1, 5]) sage: %timeit p.inverse() 10000 loops, best of 3: 67.1 µs per loop sage: p = Permutation([19, 5, 13, 8, 7, 15, 9, 10, 16, 3, 12, 6, 2, 20, 18, 11, 14, 4, 17, 1]) sage: %timeit p.inverse() 1000 loops, best of 3: 240 µs per loop sage: p = Permutation([14, 17, 1, 24, 16, 34, 19, 9, 20, 18, 36, 5, 22, 2, 27, 40, 37, 15, 3, 35, 10, 25, 21, 8, 13, 26, 12, 32, 23, 38, 11, 4, 6, 39, 31, 28, 29, 7, 30, 33]) sage: %timeit p.inverse() 1000 loops, best of 3: 857 µs per loop # AFTER sage: p = Permutation([6, 7, 8, 9, 4, 2, 3, 1, 5]) sage: %timeit p.inverse() 10000 loops, best of 3: 24.6 µs per loop sage: p = Permutation([19, 5, 13, 8, 7, 15, 9, 10, 16, 3, 12, 6, 2, 20, 18, 11, 14, 4, 17, 1]) sage: %timeit p.inverse() 10000 loops, best of 3: 41.4 µs per loop sage: p = Permutation([14, 17, 1, 24, 16, 34, 19, 9, 20, 18, 36, 5, 22, 2, 27, 40, 37, 15, 3, 35, 10, 25, 21, 8, 13, 26, 12, 32, 23, 38, 11, 4, 6, 39, 31, 28, 29, 7, 30, 33]) sage: %timeit p.inverse() 10000 loops, best of 3: 72.4 µs per loop
Updating some quirks in sage/combinat/partition.py (Andrew Mathas) #5790 --- The functions r_core(), r_quotient(), k_core(), and partition_sign() are now deprecated. These are replaced with core(), quotient(), and sign() respectively. The rewrite of the function Partition() deprecated the argument core_and_quotient. The core and the quotient can be passed as keywords of Partition().
sage: Partition(core_and_quotient=([2,1], [[2,1],[3],[1,1,1]])) /home/mvngu/.sage/temp/sage.math.washington.edu/9221/_home_mvngu__sage_init_sage_0.py:1: DeprecationWarning: "core_and_quotient=(*)" is deprecated. Use "core=[*], quotient=[*]" instead. # -*- coding: utf-8 -*- [11, 5, 5, 3, 2, 2, 2] sage: Partition(core=[2,1], quotient=[[2,1],[3],[1,1,1]]) [11, 5, 5, 3, 2, 2, 2] sage: Partition([6,3,2,2]).r_quotient(3) /home/mvngu/.sage/temp/sage.math.washington.edu/9221/_home_mvngu__sage_init_sage_0.py:1: DeprecationWarning: r_quotient is deprecated. Please use quotient instead. # -*- coding: utf-8 -*- [[], [], [2, 1]] sage: Partition([6,3,2,2]).quotient(3) [[], [], [2, 1]] sage: partition_sign([5,1,1,1,1,1]) /home/mvngu/.sage/temp/sage.math.washington.edu/9221/_home_mvngu__sage_init_sage_0.py:1: DeprecationWarning: "partition_sign deprecated. Use Partition(pi).sign() instead # -*- coding: utf-8 -*- 1 sage: Partition([5,1,1,1,1,1]).sign() 1
Cryptography
Improve S-box linear and differences matrices computation (Yann Laigle-Chapuy) #6454 --- Speed up the functions difference_distribution_matrix() and linear_approximation_matrix() of the class SBox in the module sage/crypto/mq/sbox.py. The function linear_approximation_matrix() now uses the Walsh transform. The efficiency of difference_distribution_matrix() can be up to 277x, while that for linear_approximation_matrix() can be up to 132x. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: S = mq.SR(1,4,4,8).sbox() sage: %time S.difference_distribution_matrix(); CPU times: user 77.73 s, sys: 0.00 s, total: 77.73 s Wall time: 77.73 s sage: %time S.linear_approximation_matrix(); CPU times: user 132.96 s, sys: 0.00 s, total: 132.96 s Wall time: 132.96 s # AFTER sage: S = mq.SR(1,4,4,8).sbox() sage: %time S.difference_distribution_matrix(); CPU times: user 0.28 s, sys: 0.01 s, total: 0.29 s Wall time: 0.28 s sage: %time S.linear_approximation_matrix(); CPU times: user 1.01 s, sys: 0.00 s, total: 1.01 s Wall time: 1.01 s
Elliptic Curves
Allow the method integral_points() to handle elliptic curves with large ranks (John Cremona) #6381 --- A rewrite of the method integral_x_coords_in_interval() in the class EllipticCurve_rational_field belonging to the module sage/schemes/elliptic_curves/ell_rational_field.py. The rewrite allows the method integral_points() to compute the integral points of elliptic curves with large ranks. For example, previously the following code would result in an OverflowError:
sage: D = 6611719866 sage: E = EllipticCurve([0, 0, 0, -D^2, 0]) sage: E.integral_points();
Multiplication-by-n method on elliptic curve formal groups uses the double-and-add algorithm (Hamish Ivey-Law, Tom Boothby) #6407 --- Previously, the method EllipticCurveFormalGroup.mult_by_n() was implemented by applying the group law to itself n times. However, when working over a field of characteristic zero, a faster algorithm would be used instead. The linear algorithm is now replaced with the logarithmic double-and-add algorithm, i.e. the additive version of the standard square-and-multiply algorithm. In some cases, the efficiency gain can range from 3% up to 29%. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: F = EllipticCurve(GF(101), [1, 1]).formal_group() sage: %time F.mult_by_n(100, 20); CPU times: user 0.98 s, sys: 0.00 s, total: 0.98 s Wall time: 0.98 s sage: F = EllipticCurve("37a").formal_group() sage: %time F.mult_by_n(1000000, 20); CPU times: user 0.38 s, sys: 0.00 s, total: 0.38 s Wall time: 0.38 s sage: %time F.mult_by_n(100000000, 20); CPU times: user 0.55 s, sys: 0.03 s, total: 0.58 s Wall time: 0.58 s # AFTER sage: F = EllipticCurve(GF(101), [1, 1]).formal_group() sage: %time F.mult_by_n(100, 20); CPU times: user 0.96 s, sys: 0.00 s, total: 0.96 s Wall time: 0.95 s sage: F = EllipticCurve("37a").formal_group() sage: %time F.mult_by_n(1000000, 20); CPU times: user 0.44 s, sys: 0.01 s, total: 0.45 s Wall time: 0.45 s sage: %time F.mult_by_n(100000000, 20); CPU times: user 0.40 s, sys: 0.01 s, total: 0.41 s Wall time: 0.41 s
Graphics
Plotting 3-D Bezier paths (Emily Kirkman) #6098 --- New function bezier3d() for plotting a 3-dimensional Bezier path. Here are some examples for working with this function:
sage: bezier3d([[(0,0,0), (1,0,0), (0,1,0), (0,1,1)]]).show(zoom=1.2)
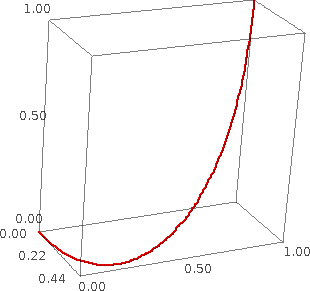
sage: path = [[(0,0,0),(.5,.1,.2),(.75,3,-1),(1,1,0)],[(.5,1,.2),(1,.5,0)],[(.7,.2,.5)]] sage: bezier3d(path, color='green').show(zoom=1.2)
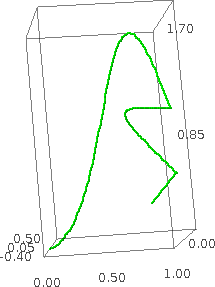
Passing extra options to show() (Bill Cauchois) #5651 --- Extra optional arguments to plotting functions can now be passed on to the function show(). This passing of optional arguments is implemented for the following plotting modules:
sage/plot/arrow.py
sage/plot/bar_chart.py
sage/plot/bezier_path.py
sage/plot/circle.py
sage/plot/complex_plot.pyx
sage/plot/contour_plot.py
sage/plot/density_plot.py
sage/plot/disk.py
sage/plot/line.py
sage/plot/matrix_plot.py
sage/plot/plot.py
sage/plot/plot_field.py
sage/plot/point.py
sage/plot/polygon.py
sage/plot/scatter_plot.py
sage/plot/text.py Each of the following examples demonstrates equivalent code to obtain a plot:
sage: arrow((-2, 2), (7,1), frame=True) sage: arrow((-2, 2), (7,1)).show(frame=True)
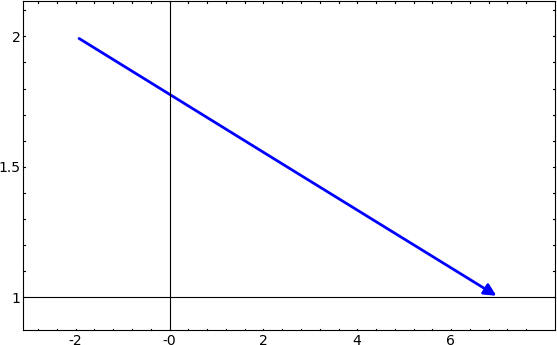
sage: bar_chart([-2,8,-7,3], rgbcolor=(1,0,0), axes=False) sage: bar_chart([-2,8,-7,3], rgbcolor=(1,0,0)).show(axes=False)
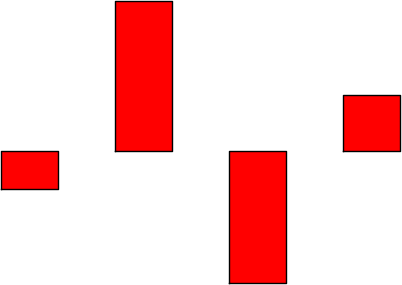
sage: bezier_path([[(0,1),(.5,0),(1,1)]], fontsize=20) sage: bezier_path([[(0,1),(.5,0),(1,1)]]).show(fontsize=20)
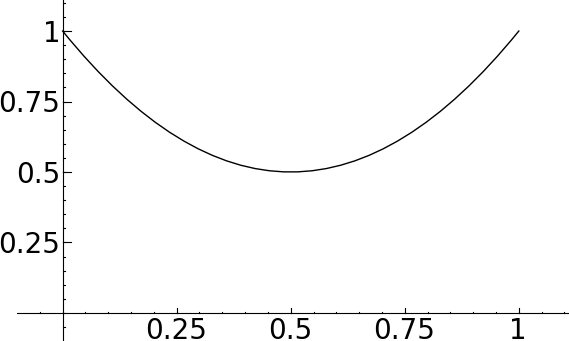
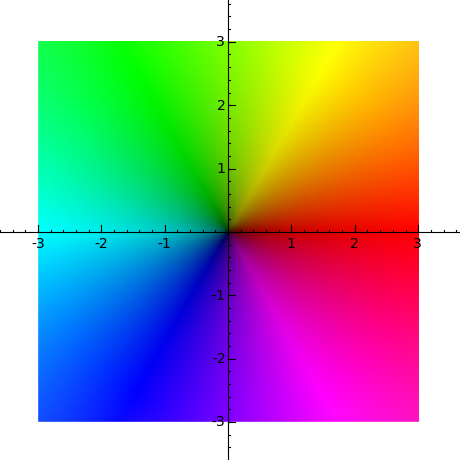
sage: complex_plot(lambda z: z, (-3, 3), (-3, 3), figsize=[5,5]) sage: complex_plot(lambda z: z, (-3, 3), (-3, 3)).show(figsize=[5,5])
Graph Theory
Cliquer as a standard package (Nathann Cohen) #6355 --- Cliquer is a set of C routines for finding cliques in an arbitrary weighted graph. It uses an exact branch-and-bound algorithm recently developed by Patric Ostergard and mainly written by Sampo Niskanen. It is published under the GPL license. Here are some examples for working with the new cliquer spkg:
sage: max_clique(graphs.PetersenGraph()) [7, 9] sage: all_max_clique(graphs.PetersenGraph()) [[2, 7], [7, 9], [6, 8], [6, 9], [0, 4], [4, 9], [5, 7], [0, 5], [5, 8], [3, 4], [2, 3], [3, 8], [1, 6], [0, 1], [1, 2]] sage: clique_number(Graph("DJ{")) 4 sage: clique_number(Graph({0:[1,2,3], 1:[2], 3:[0,1]})) 3 sage: list_composition([1,3,'a'], {'a':'b', 1:2, 2:3, 3:4}) [2, 4, 'b']
Faster algorithm to compute maximum cliques (Nathann Cohen) #5793 --- With the inclusion of cliquer as a standard spkg, the following functions can now use the cliquer algorithm:
Graph.max_clique() --- Returns the vertex set of a maximum complete subgraph.
Graph.cliques_maximum() --- Returns the list of all maximum cliques, with each clique represented by a list of vertices. A clique is an induced complete subgraph and a maximal clique is one of maximal order.
Graph.clique_number() --- Returns the size of the largest clique of the graph.
Graph.cliques_vertex_clique_number() --- Returns a list of sizes of the largest maximal cliques containing each vertex. This returns a single value if there is only one input vertex.
Graph.independent_set() --- Returns a maximal independent set, which is a set of vertices which induces an empty subgraph.
These functions already exist in Sage. Cliquer does not bring to Sage any new feature, but a huge efficiency improvement in computing clique numbers. The NetworkX 0.36 algorithm is very slow in its computation of these functions, even though it remains faster than cliquer for the computation of Graph.cliques_vertex_clique_number(). The algorithms in the cliquer spkg scale very well as the number of vertices in a graph increases. Here is a comparison between the implementation of NetworkX 0.36 and cliquer on computing the clique number of a graph. Timing statistics were obtained using the machine sage.math:
sage: g = graphs.RandomGNP(100, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 0.64 s, sys: 0.01 s, total: 0.65 s Wall time: 0.65 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 0.02 s, sys: 0.00 s, total: 0.02 s Wall time: 0.02 s sage: g = graphs.RandomGNP(200, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 9.68 s, sys: 0.01 s, total: 9.69 s Wall time: 9.68 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 0.09 s, sys: 0.00 s, total: 0.09 s Wall time: 0.09 s sage: g = graphs.RandomGNP(300, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 69.98 s, sys: 0.10 s, total: 70.08 s Wall time: 70.09 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 0.23 s, sys: 0.00 s, total: 0.23 s Wall time: 0.23 s sage: g = graphs.RandomGNP(400, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 299.32 s, sys: 0.29 s, total: 299.61 s Wall time: 299.64 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 0.54 s, sys: 0.00 s, total: 0.54 s Wall time: 0.53 s sage: g = graphs.RandomGNP(500, 0.4) sage: %time g.clique_number(algorithm="networkx"); CPU times: user 1178.85 s, sys: 1.30 s, total: 1180.15 s Wall time: 1180.16 s sage: %time g.clique_number(algorithm="cliquer"); CPU times: user 1.09 s, sys: 0.00 s, total: 1.09 s Wall time: 1.09 s
Support the syntax g.add_edge((u,v), label=l) for C graphs (Robert Miller) #6540 --- The following syntax is supported. However, note that the label keyword must be used:
sage: G = Graph() sage: G.add_edge((1,2), label="my label") sage: G.edges() [(1, 2, 'my label')] sage: G = Graph() sage: G.add_edge((1,2), "label") sage: G.edges() [((1, 2), 'label', None)]
Fast subgraphs by building the graph instead of deleting things (Jason Grout) #6578 --- Subgraphs can now be constructed by building a new graph from a number of vertices and edges. This is in contrast to the previous default algorithm where subgraphs were constructed by deleting edges and vertices. In some cases, the efficiency gain of the new subgraph construction implementation can be up to 17x. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: g = graphs.PathGraph(Integer(10e4)) sage: %time g.subgraph(range(20)); CPU times: user 1.89 s, sys: 0.03 s, total: 1.92 s Wall time: 1.92 s sage: g = graphs.PathGraph(Integer(10e4) * 5) sage: %time g.subgraph(range(20)); CPU times: user 14.92 s, sys: 0.05 s, total: 14.97 s Wall time: 14.97 s sage: g = graphs.PathGraph(Integer(10e5)) sage: %time g.subgraph(range(20)); CPU times: user 47.77 s, sys: 0.29 s, total: 48.06 s Wall time: 48.06 s # AFTER sage: g = graphs.PathGraph(Integer(10e4)) sage: %time g.subgraph(range(20)); CPU times: user 0.27 s, sys: 0.01 s, total: 0.28 s Wall time: 0.28 s sage: g = graphs.PathGraph(Integer(10e4) * 5) sage: %time g.subgraph(range(20)); CPU times: user 1.34 s, sys: 0.03 s, total: 1.37 s Wall time: 1.37 s sage: g = graphs.PathGraph(Integer(10e5)) sage: %time g.subgraph(range(20)); CPU times: user 2.66 s, sys: 0.04 s, total: 2.70 s Wall time: 2.70 s
Interfaces
Magma interface: make magma_colon_equals() mode work in both command line and notebook (William Stein) #6395 --- Exposes the magma_colon_equals() option in the notebook. For example, one can now do the following in the notebook:
sage: magma._preparse_colon_equals = False sage: magma._preparse('a = 5') 'a = 5' sage: magma._preparse_colon_equals = True sage: magma._preparse('a = 5') 'a := 5' sage: magma._preparse('a = 5; b := 7; c =a+b;') 'a := 5; b := 7; c :=a+b;'
Viewing a Sage object with view(object, viewer='pdf') (Nicolas M. Thiéry) #6591 --- Typical uses include:
- you prefer your PDF browser to your DVI browser
- you want to view LaTeX snippets which are not displayed well in DVI viewers or jsMath, e.g. images produced by tikzpicture.
Linear Algebra
Make NumPy play nice with Sage types (Robert Bradshaw) #5081 --- This improves data conversion between NumPy and Sage. For example, one can now do this:
And this:sage: from scipy import stats sage: stats.uniform(0,15).ppf([0.5,0.7]) array([ 7.5, 10.5])
sage: from scipy import * sage: from pylab import * sage: sample_rate = 1000.0 sage: t = r_[0:0.6:1/sample_rate] sage: N = len(t) sage: s = [sin(2*pi*50*elem) + sin(2*pi*70*elem + (pi/4)) for elem in t] sage: S = fft(s) sage: f = sample_rate*r_[0:(N/2)] / N sage: n = len(f) sage: line(zip(f, abs(S[0:n]) / N))
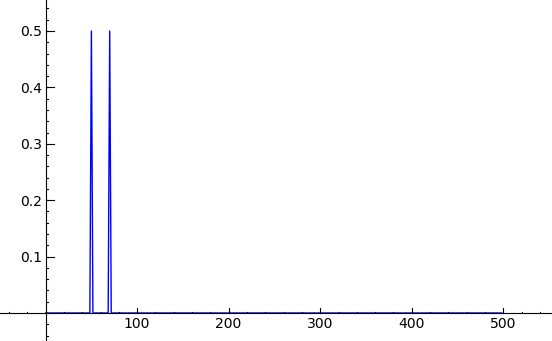
Fast slicing of sparse matrices (Jason Grout) #6553 --- The efficiency gain for slicing sparse matrices can range from 10x up to 147x. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: A = random_matrix(ZZ, 100000, density=0.00005, sparse=True) sage: %time A[50000:,:50000]; CPU times: user 298.84 s, sys: 0.05 s, total: 298.89 s Wall time: 298.95 s sage: A = random_matrix(ZZ, 10000, density=0.00005, sparse=True) sage: %time A[5000:,:5000]; CPU times: user 2.50 s, sys: 0.00 s, total: 2.50 s Wall time: 2.50 s # AFTER sage: A = random_matrix(ZZ, 100000, density=0.00005, sparse=True) sage: %time A[50000:,:50000]; CPU times: user 1.91 s, sys: 0.09 s, total: 2.00 s Wall time: 2.02 s sage: A = random_matrix(ZZ, 10000, density=0.00005, sparse=True) sage: %time A[5000:,:5000]; CPU times: user 0.23 s, sys: 0.00 s, total: 0.23 s Wall time: 0.24 s
Plotting sparse matrices efficiently (Jason Grout) #6554 --- Previously, plotting a sparse matrix would convert the matrix to a dense matrix, resulting in the whole process taking increasingly longer time as the dimensions of the matrix increase. Where a matrix is sparse, the matrix plotting function now uses SciPy's sparse matrix functionalities, which can handle large matrices. In some cases, the performance improvement can range from 380x up to 98000x. The following timing statistics were obtained using the machine mod.math:
# BEFORE sage: A = random_matrix(ZZ, 5000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 60.25 s, sys: 0.69 s, total: 60.94 s Wall time: 60.94 s sage: A = random_matrix(ZZ, 10000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 241.31 s, sys: 3.03 s, total: 244.34 s Wall time: 244.35 s sage: A = random_matrix(ZZ, 15000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 544.02 s, sys: 6.85 s, total: 550.87 s Wall time: 550.86 s sage: A = random_matrix(ZZ, 20000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 972.85 s, sys: 13.36 s, total: 986.21 s Wall time: 986.21 s # AFTER sage: A = random_matrix(ZZ, 5000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.05 s, sys: 0.04 s, total: 0.09 s Wall time: 0.16 s sage: A = random_matrix(ZZ, 10000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s sage: A = random_matrix(ZZ, 15000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s sage: A = random_matrix(ZZ, 20000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s
In Sage 4.1, the following would quickly consume gigabytes of RAM on a system and may result in a MemoryError:
sage: A = random_matrix(ZZ, 100000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 0.63 s, sys: 0.01 s, total: 0.64 s Wall time: 0.63 s sage: A = random_matrix(ZZ, 1000000, density=0.00001, sparse=True) sage: %time matrix_plot(A, marker=','); CPU times: user 1933.41 s, sys: 2.97 s, total: 1936.38 s Wall time: 1937.31 s
Elementwise (Hadamard) product of matrices (Rob Beezer) #6301 --- Given matrices A and B of the same size, C = A.elementwise_product(B) creates the new matrix C, of the same size, with entries given by C[i,j] = A[i,j] * B[i,j]. The multiplication occurs in a ring defined by Sage's coercion model, as appropriate for the base rings of A and B (or an error is raised if there is no sensible common ring). In particular, if A and B are defined over a noncommutative ring, the operation is also noncommutative. The implementation is different for dense matrices versus sparse matrices, but there are probably further optimizations available for specific rings. This operation is often called the Hadamard product. Here is an example on using elementwise matrix product:
sage: G = matrix(GF(3), 2, [0,1,2,2]) sage: H = matrix(ZZ, 2, [1,2,3,4]) sage: J = G.elementwise_product(H); J [0 2] [0 2] sage: J.parent() Full MatrixSpace of 2 by 2 dense matrices over Finite Field of size 3
Modular Forms
Efficient implementation of index for Gamma(N) (Simon Morris) #6606 --- The new implementation provides massive speed-up for the function Gamma(N). The following statistics were obtained using the machine mod.math:
# BEFORE sage: %time [Gamma(n).index() for n in [1..19]]; CPU times: user 14369.53 s, sys: 75.18 s, total: 14444.71 s Wall time: 14445.22 s sage: %time Gamma(32041).index(); # hangs for hours # AFTER sage: %time [Gamma(n).index() for n in [1..19]]; CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s Wall time: 0.00 s sage: timeit("[Gamma(n).index() for n in [1..19]]") 125 loops, best of 3: 2.27 ms per loop sage: %time Gamma(32041).index(); CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s sage: %timeit Gamma(32041).index() 10000 loops, best of 3: 110 µs per loop
Weight 1 Eisenstein series (David Loeffler) #6071 --- Add support for computing weight 1 Eisenstein series. Here's an example:
sage: M = ModularForms(DirichletGroup(13).0, 1) sage: M.eisenstein_series() [ -1/13*zeta12^3 + 6/13*zeta12^2 + 4/13*zeta12 + 2/13 + q + (zeta12 + 1)*q^2 + zeta12^2*q^3 + (zeta12^2 + zeta12 + 1)*q^4 + (-zeta12^3 + 1)*q^5 + O(q^6) ]
Efficient calculation of Jacobi sums (David Loeffler) #6534 --- For small primes, calculating Jacobi sums using the definition directly can result in an efficiency improvement of up to 624x. The following timing statistics were obtained using the machine mod.math:
There is also support for computing a Jacobi sum with values in a finite field:# BEFORE sage: chi = DirichletGroup(67).0 sage: psi = chi**3 sage: time chi.jacobi_sum(psi); CPU times: user 6.24 s, sys: 0.00 s, total: 6.24 s Wall time: 6.24 s # AFTER sage: chi = DirichletGroup(67).0 sage: psi = chi**3 sage: time chi.jacobi_sum(psi); CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s
sage: g = DirichletGroup(17, GF(9,'a')).0 sage: g.jacobi_sum(g**2) 2*a
Notebook
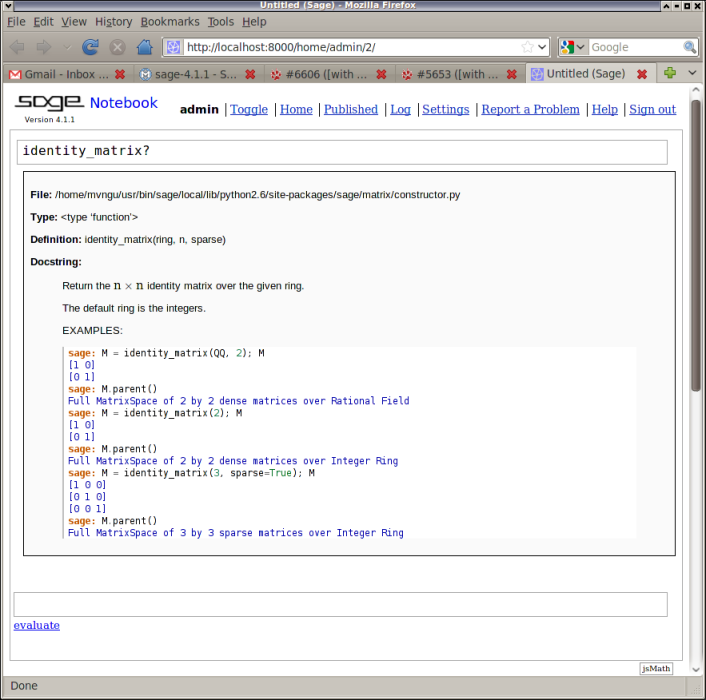
Display docstrings in the notebook using HTML and jsMath (Tom Boothby, Evan Fosmark, John Palmieri, Mitesh Patel) #5653 --- When viewing docstrings using the notebook, these are presented using HTML and jsMath. For example, if one does any of the following
identity_matrix([TAB] identity_matrix?[SHIFT-RETURN] identity_matrix?[TAB]
then the docstring for the function identity_matrix() would be presented as in this figure:
Number Theory
Intersection of ideals in a number field (David Loeffler) #6457 --- New function intersection() of the class NumberFieldIdeal in the module sage/rings/number_field/number_field_ideal.py for computing the ideals in a number field. Here are some examples on using intersection():
sage: K.<a> = QuadraticField(-11) sage: p = K.ideal((a + 1)/2); q = K.ideal((a + 3)/2) sage: p.intersection(q) == q.intersection(p) == K.ideal(a-2) True sage: # An example with non-principal ideals sage: L.<a> = NumberField(x^3 - 7) sage: p = L.ideal(a^2 + a + 1, 2) sage: q = L.ideal(a+1) sage: p.intersection(q) == L.ideal(8, 2*a + 2) True sage: # A relative example sage: L.<a,b> = NumberField([x^2 + 11, x^2 - 5]) sage: A = L.ideal([15, (-3/2*b + 7/2)*a - 8]) sage: B = L.ideal([6, (-1/2*b + 1)*a - b - 5/2]) sage: A.intersection(B) == L.ideal(-1/2*a - 3/2*b - 1) True
Computing Heegner points (Robert Bradshaw) #6045 --- Adds a Heegner point method to elliptic curves over QQ. The new method is heegner_point() which can be found in the class EllipticCurve_rational_field of the module sage/schemes/elliptic_curves/ell_rational_field.py. Here are some examples on using heegner_point():
sage: E = EllipticCurve('37a') sage: E.heegner_discriminants_list(10) [-7, -11, -40, -47, -67, -71, -83, -84, -95, -104] sage: E.heegner_point(-7) (0 : 0 : 1) sage: P = E.heegner_point(-40); P (a : -a + 1 : 1) sage: P = E.heegner_point(-47); P (a : -a^4 - a : 1) sage: P[0].parent() Number Field in a with defining polynomial x^5 - x^4 + x^3 + x^2 - 2*x + 1 sage: # Working out the details manually sage: P = E.heegner_point(-47, prec=200) sage: f = algdep(P[0], 5); f x^5 - x^4 + x^3 + x^2 - 2*x + 1 sage: f.discriminant().factor() 47^2
Support primes_of_degree_one() for relative extensions (David Loeffler) #6396 --- For example, one can now do this:
sage: N.<a,b> = NumberField([x^2 + 1, x^2 - 5]) sage: ids = N.primes_of_degree_one_list(10); ids [Fractional ideal (2*a + 1/2*b - 1/2), Fractional ideal ((-1/2*b - 1/2)*a - b), Fractional ideal (b*a + 1/2*b + 3/2), Fractional ideal ((-1/2*b - 3/2)*a + b - 1), Fractional ideal ((-b + 1)*a + b), Fractional ideal (3*a + 1/2*b - 1/2), Fractional ideal ((1/2*b - 1/2)*a + 3/2*b - 1/2), Fractional ideal ((-1/2*b - 5/2)*a - b + 1), Fractional ideal (2*a - 3/2*b - 1/2), Fractional ideal (3*a + 1/2*b + 5/2)] sage: [x.absolute_norm() for x in ids] [29, 41, 61, 89, 101, 109, 149, 181, 229, 241] sage: ids[9] == N.ideal(3*a + 1/2*b + 5/2) True
Inverse modulo an ideal in a relative number field (David Loeffler) #6458 --- Support for computing the inverse modulo an ideal in the ring of integers of a relative field. Here's an example:
sage: K.<a,b> = NumberField([x^2 + 1, x^2 - 3]) sage: I = K.ideal(17*b - 3*a) sage: x = I.integral_basis(); x [438, -b*a + 309, 219*a - 219*b, 156*a - 154*b] sage: V, _, phi = K.absolute_vector_space() sage: V.span([phi(u) for u in x], ZZ) == I.free_module() True
Numerical
Further NumPy type conversions (Robert Bradshaw, Jason Grout) #6506 --- Support improved handling of ZZ, QQ, and CC between NumPy and Sage. Here are some examples:
sage: import numpy sage: numpy.array([1.0, 2.5j]) array([ 1.+0.j , 0.+2.5j]) sage: numpy.array([1.0, 2.5j]).dtype dtype('complex128') sage: numpy.array([1.000000000000000000000000000000000000j]).dtype dtype('object') sage: numpy.array([1, 2, 3]) array([1, 2, 3]) sage: numpy.array([1, 2, 3]).dtype dtype('int64') sage: numpy.array(2**40).dtype dtype('int64') sage: numpy.array(2**400).dtype dtype('object') sage: numpy.array([1,2,3,0.1]).dtype dtype('float64') sage: numpy.array(QQ(2**40)).dtype dtype('int64') sage: numpy.array(QQ(2**400)).dtype dtype('object') sage: numpy.array([1, 1/2, 3/4]) array([ 1. , 0.5 , 0.75])
Packages
Update the ATLAS spkg to version 3.8.3.p7 (David Kirkby, Minh Van Nguyen) #6558 #6738 --- This adds support for building ATLAS under Solaris on a Sun SPARC processor.
Update the NTL spkg to version 5.4.2.p9 (David Kirkby) #6380 --- This adds support for building NTL under Solaris on a Sun SPARC processor.
Update the zn_poly spkg to version 0.9.p1 (David Kirkby) #6443 --- This adds support for building zn_poly under Solaris on a Sun SPARC processor.
Update the Pari/GP spkg to version 2.3.3.p1 (David Kirkby) #6445 --- This adds support for building Pari under Solaris on a Sun SPARC processor.
Update the FLINT spkg to version 1.3.0.p2 (David Kirkby) #6451 --- This fixes a Solaris specific bug in FLINT.
Update the MPFR spkg to version 2.4.1.p0 (Paul Zimmermann, David Kirkby) #6453 --- This fixes a number of test failures under Solaris on a Sun SPARC processor.
Update the PolyBoRi spkg to version 0.5rc.p9 (David Kirkby) #6528 --- This fixes a Solaris specific bug in compiling PolyBoRi with the Sun compiler.
Upgrade tinyMCE to version 3.2.4.1 upstream release (Jason Grout) #6143 --- This version of tinyMCE has many fixes for Safari and a greatly improved paste-from-word functionality.
Upgrade Cython to version 0.11.2.1 latest upstream release (Robert Bradshaw) #6438.
Update the NumPy spkg to version 1.3.0.p1 (William Stein) #6493 --- This fixes a bug in compiling NumPy under 64-bit OS X.
Upgrade Singular to version singular-3-1-0-2-20090620.p0 (David Kirkby) #6563 --- This fixes a Solaris specific bug when compiling Singular under Solaris on a Sun SPARC processor.
New optional spkg GLPK version 4.38 (Nathann Cohen) #6602 --- GLPK is a linear program solver that can also solve mixed integer programs.
New optional spkg OpenOpt version 0.24 (William Stein) #6302 --- OpenOpt is a numerical optimization with various solvers.
New optional package p_group_cohomology version 1.0.2 (Simon A. King, David J. Green) #6491 --- The package p_group_cohomology can compute the cohomology ring of a group with coefficients in a finite field of order p. Its features include:
- Compute the cohomology ring with coefficients in GF(p) for any finite p-group, in terms of a minimal generating set and a minimal set of algebraic relations. We use Benson's criterion to prove the completeness of the ring structure.
- Compute depth, dimension, Poincare series and a-invariants of the cohomology rings.
- Compute the nil radical
- Construct induced homomorphisms.
- The package includes a list of cohomology rings for all groups of order 64.
- With the package, the cohomology for all groups of order 128 and for the Sylow 2-subgroup of the third Conway group (order 1024) was computed for the first time. The result of these and many other computations (e.g., all but 6 groups of order 243) is accessible in a repository on sage.math.
- Data that are included with the package:
sage: from pGroupCohomology import CohomologyRing sage: H = CohomologyRing(64,132) # this is included in the package, hence, the ring structure is already there sage: print H Cohomology ring of Small Group number 132 of order 64 with coefficients in GF(2) Computation complete Minimal list of generators: [a_2_4, a 2-Cochain in H^*(SmallGroup(64,132); GF(2)), c_2_5, a 2-Cochain in H^*(SmallGroup(64,132); GF(2)), c_4_12, a 4-Cochain in H^*(SmallGroup(64,132); GF(2)), a_1_0, a 1-Cochain in H^*(SmallGroup(64,132); GF(2)), a_1_1, a 1-Cochain in H^*(SmallGroup(64,132); GF(2)), b_1_2, a 1-Cochain in H^*(SmallGroup(64,132); GF(2))] Minimal list of algebraic relations: [a_1_0*a_1_1, a_1_0*b_1_2, a_1_1^3+a_1_0^3, a_2_4*a_1_0, a_1_1^2*b_1_2^2+a_2_4*a_1_1*b_1_2+a_2_4^2+c_2_5*a_1_1^2] sage: H.depth() 2 sage: H.a_invariants() [-Infinity, -Infinity, -3, -3] sage: H.poincare_series() (-t^2 - t - 1)/(t^6 - 2*t^5 + t^4 - t^2 + 2*t - 1) sage: H.nil_radical() a_1_0, a_1_1, a_2_4
- Data from the repository on sage.math:
sage: H = CohomologyRing(128,562) # if there is internet connection, the ring data are downloaded behind the scenes sage: len(H.gens()) 35 sage: len(H.rels()) 486 sage: H.depth() 1 sage: H.a_invariants() [-Infinity, -4, -3, -3] sage: H.poincare_series() (t^14 - 2*t^13 + 2*t^12 - t^11 - t^10 + t^9 - 2*t^8 + 2*t^7 - 2*t^6 + 2*t^5 - 2*t^4 + t^3 - t^2 - 1)/(t^17 - 3*t^16 + 4*t^15 - 4*t^14 + 4*t^13 - 4*t^12 + 4*t^11 - 4*t^10 + 4*t^9 - 4*t^8 + 4*t^7 - 4*t^6 + 4*t^5 - 4*t^4 + 4*t^3 - 4*t^2 + 3*t - 1)
- Some computation from scratch, involving different ring presentations and induced maps:
sage: tmp_root = tmp_filename() sage: CohomologyRing.set_user_db(tmp_root) sage: H0 = CohomologyRing.user_db(8,3,websource=False) sage: print H0 Cohomology ring of Dihedral group of order 8 with coefficients in GF(2) Computed up to degree 0 Minimal list of generators: [] Minimal list of algebraic relations: [] sage: H0.make() sage: print H0 Cohomology ring of Dihedral group of order 8 with coefficients in GF(2) Computation complete Minimal list of generators: [c_2_2, a 2-Cochain in H^*(D8; GF(2)), b_1_0, a 1-Cochain in H^*(D8; GF(2)), b_1_1, a 1-Cochain in H^*(D8; GF(2))] Minimal list of algebraic relations: [b_1_0*b_1_1] sage: G = gap('DihedralGroup(8)') sage: H1 = CohomologyRing.user_db(G,GroupName='GapD8',websource=False) sage: H1.make() sage: print H1 # the ring presentation is different ... Cohomology ring of GapD8 with coefficients in GF(2) Computation complete Minimal list of generators: [c_2_2, a 2-Cochain in H^*(GapD8; GF(2)), b_1_0, a 1-Cochain in H^*(GapD8; GF(2)), b_1_1, a 1-Cochain in H^*(GapD8; GF(2))] Minimal list of algebraic relations: [b_1_1^2+b_1_0*b_1_1] sage: phi = G.IsomorphismGroups(H0.group()) sage: phi_star = H0.hom(phi,H1) sage: phi_star_inv = phi_star^(-1) # ... but the rings are isomorphic sage: [X==phi_star_inv(phi_star(X)) for X in H0.gens()] [True, True, True, True] sage: [X==phi_star(phi_star_inv(X)) for X in H1.gens()] [True, True, True, True]
- An example with an odd prime:
sage: H = CohomologyRing(81,8) # this needs to be computed from scratch sage: H.make() sage: H.gens() [1, a_2_1, a 2-Cochain in H^*(SmallGroup(81,8); GF(3)), a_2_2, a 2-Cochain in H^*(SmallGroup(81,8); GF(3)), b_2_0, a 2-Cochain in H^*(SmallGroup(81,8); GF(3)), a_4_1, a 4-Cochain in H^*(SmallGroup(81,8); GF(3)), b_4_2, a 4-Cochain in H^*(SmallGroup(81,8); GF(3)), b_6_3, a 6-Cochain in H^*(SmallGroup(81,8); GF(3)), c_6_4, a 6-Cochain in H^*(SmallGroup(81,8); GF(3)), a_1_0, a 1-Cochain in H^*(SmallGroup(81,8); GF(3)), a_1_1, a 1-Cochain in H^*(SmallGroup(81,8); GF(3)), a_3_2, a 3-Cochain in H^*(SmallGroup(81,8); GF(3)), a_5_2, a 5-Cochain in H^*(SmallGroup(81,8); GF(3)), a_5_3, a 5-Cochain in H^*(SmallGroup(81,8); GF(3)), a_7_5, a 7-Cochain in H^*(SmallGroup(81,8); GF(3))] sage: len(H.rels()) 59 sage: H.depth() 1 sage: H.a_invariants() [-Infinity, -3, -2] sage: H.poincare_series() (t^4 - t^3 + t^2 + 1)/(t^6 - 2*t^5 + 2*t^4 - 2*t^3 + 2*t^2 - 2*t + 1) sage: H.nil_radical() a_1_0, a_1_1, a_2_1, a_2_2, a_3_2, a_4_1, a_5_2, a_5_3, b_2_0*b_4_2, a_7_5, b_2_0*b_6_3, b_6_3^2+b_4_2^3
