|
Size: 17526
Comment: corrected authorship of E6 patch
|
Size: 30730
Comment: Add link to nicely formatted release tour
|
| Deletions are marked like this. | Additions are marked like this. |
| Line 3: | Line 3: |
| Sage 4.0 was released on FIXME. For the official, comprehensive release note, please refer to [[http://www.sagemath.org/src/announce/sage-4.0.txt|sage-4.0.txt]]. A nicely formatted version of this release tour can be found at FIXME. The following points are some of the foci of this release: * |
Sage 4.0 was released on May 29, 2009. For the official, comprehensive release note, please refer to [[http://sage.math.washington.edu/home/mhansen/sage-4-release-notes|sage-4.0.txt]]. A nicely formatted version of this release tour can be found at [[http://mvngu.wordpress.com/2009/06/03/sage-4-0-released/|this Wordpress blog]]. The following points are some of the foci of this release: * New symbolics based on Pynac * Bring doctest coverage up to at least 75% * Solaris 10 support (at least for gcc 4.3.x + gmake) * Switch from Clisp to ECL * gcc 4.4.0 support * OS X 64-bit support |
| Line 46: | Line 51: |
* FIXME: summarize #6036 |
|
| Line 73: | Line 76: |
| == Build == == Calculus == |
|
| Line 82: | Line 80: |
| * FIXME: summarize #5582 | * Coercion from float to rationals (Robert Bradshaw) -- One can now coerce a number of type float to {{{QQ}}}. Here's an example: {{{ sage: a = float(1.0) sage: QQ(a) 1 sage: type(a); type(QQ(a)) <type 'float'> <type 'sage.rings.rational.Rational'> }}} |
| Line 112: | Line 118: |
| * Crystal of letters for type E (Brant Jones and Anne Schilling) -- Support crystal of letters for type E corresponding to the highest weight crystal {{{B(\Lambda_1)}}} and its dual {{{B(\Lambda_6)}}} (using the Sage labeling convention of the Dynkin nodes). Here are some examples: | * Crystal of letters for type E (Brant Jones, Anne Schilling) -- Support crystal of letters for type E corresponding to the highest weight crystal {{{B(\Lambda_1)}}} and its dual {{{B(\Lambda_6)}}} (using the Sage labeling convention of the Dynkin nodes). Here are some examples: |
| Line 210: | Line 216: |
| Furthermore, the new module {{{sage/rings/polynomial/symmetric_ideal.py}}} supports ideals of polynomial rings in a countably infinite number of variables that are invariant under variable permuation. Symmetric reduction of infinite polynomials is provided by the new module {{{sage/rings/polynomial/symmetric_reduction.pyx}}}. == Distribution == == Doctest == == Documentation == |
Furthermore, the new module {{{sage/rings/polynomial/symmetric_ideal.py}}} supports ideals of polynomial rings in a countably infinite number of variables that are invariant under variable permutation. Symmetric reduction of infinite polynomials is provided by the new module {{{sage/rings/polynomial/symmetric_reduction.pyx}}}. |
| Line 265: | Line 262: |
| * FIXME: summarize #6066 * FIXME: summarize #3932 * FIXME: summarize #5940 * FIXME: summarize #6086 |
* Optimize the construction of large Sage graphs (Radoslav Kirov) -- The construction of large Sage graphs is now up to 19x faster than previously. The following timing statistics were obtained using the machine sage.math: {{{ # BEFORE sage: D = {} sage: for i in xrange(10^3): ....: D[i] = [i+1, i-1] ....: sage: timeit("g = Graph(D)") 5 loops, best of 3: 1.02 s per loop # AFTER sage: D = {} sage: for i in xrange(10^3): ....: D[i] = [i+1, i-1] ....: sage: timeit("g = Graph(D)") 5 loops, best of 3: 51.2 ms per loop }}} * Generate size {{{n}}} trees in linear time (Ryan Dingman) -- The speed-up can be up to 3400x. However, the efficiency gain is greater as {{{n}}} becomes larger. The following timing statistics were produced using the machine sage.math: {{{ # BEFORE sage: %time L = list(graphs.trees(2)) CPU times: user 0.13 s, sys: 0.02 s, total: 0.15 s Wall time: 0.18 s sage: %time L = list(graphs.trees(4)) CPU times: user 0.02 s, sys: 0.00 s, total: 0.02 s Wall time: 0.02 s sage: %time L = list(graphs.trees(6)) CPU times: user 0.08 s, sys: 0.00 s, total: 0.08 s Wall time: 0.07 s sage: %time L = list(graphs.trees(8)) CPU times: user 0.59 s, sys: 0.00 s, total: 0.59 s Wall time: 0.60 s sage: %time L = list(graphs.trees(10)) CPU times: user 34.48 s, sys: 0.02 s, total: 34.50 s Wall time: 34.51 s # AFTER sage: %time L = list(graphs.trees(2)) CPU times: user 0.11 s, sys: 0.02 s, total: 0.13 s Wall time: 0.15 s sage: %time L = list(graphs.trees(4)) CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.00 s sage: %time L = list(graphs.trees(6)) CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s Wall time: 0.00 s sage: %time L = list(graphs.trees(8)) CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s Wall time: 0.00 s sage: %time L = list(graphs.trees(10)) CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s sage: %time L = list(graphs.trees(12)) CPU times: user 0.06 s, sys: 0.00 s, total: 0.06 s Wall time: 0.05 s sage: %time L = list(graphs.trees(14)) CPU times: user 0.51 s, sys: 0.01 s, total: 0.52 s Wall time: 0.52 s }}} |
| Line 277: | Line 334: |
| * FIXME: summarize #5249 * FIXME: summarize #4875 |
* Implicit Surfaces (Bill Cauchois, Carl Witty) -- New function {{{implicit_plot3d}}} for plotting level sets of 3-D functions. The documentation contains many examples. Here's a sphere contained inside a tube-like sphere: {{{ sage: x, y, z = var("x, y, z") sage: T = 1.61803398875 sage: p = 2 - (cos(x + T*y) + cos(x - T*y) + cos(y + T*z) + cos(y - T*z) + cos(z - T*x) + cos(z + T*x)) sage: r = 4.77 sage: implicit_plot3d(p, (-r, r), (-r, r), (-r, r), plot_points=40, zoom=1.2).show() }}} {{attachment:sphere-inside-tube.png}} Here's a Klein bottle: {{{ sage: x, y, z = var("x, y, z") sage: implicit_plot3d((x^2+y^2+z^2+2*y-1)*((x^2+y^2+z^2-2*y-1)^2-8*z^2)+16*x*z*(x^2+y^2+z^2-2*y-1), (x, -3, 3), (y, -3.1, 3.1), (z, -4, 4), zoom=1.2) }}} {{attachment:klein-bottle.png}} This example shows something resembling a water droplet: {{{ sage: x, y, z = var("x, y, z") sage: implicit_plot3d(x^2 +y^2 -(1-z)*z^2, (x, -1.5, 1.5), (y, -1.5, 1.5), (z, -1, 1), zoom=1.2) }}} {{attachment:water-droplet.png}} * Fixed bug in rendering 2D polytopes embedded in 3D (Arnauld Bergeron, Bill Cauchois, Marshall Hampton). |
| Line 395: | Line 473: |
| * Refactor matrix kernels (Rob Beezer) -- The core section of kernel computation for each (specialized) class is now moved into the method {{{right_kernel()}}}. Mostly these would replace {{{kernel()}}} methods that are computing left kernels. A call to {{{kernel()}}} or {{{left_kernel()}}} should arrive at the top of the hierarchy where it would take a transpose and call the (specialized) {{{right_kernel()}}}. So there wouldn't be a change in behavior in routines currently calling {{{kernel()}}} or {{{left_kernel()}}}, and Sage's preference for the left is retained by having the vanilla {{{kernel()}}} give back a left kernel. The speed-up for the computation of left kernels is up to 5% faster, and the computation of right kernels is up to 31% by eliminating paired transposes. The followingn timing statistics were obtained using sage.math: | * Refactor matrix kernels (Rob Beezer) -- The core section of kernel computation for each (specialized) class is now moved into the method {{{right_kernel()}}}. Mostly these would replace {{{kernel()}}} methods that are computing left kernels. A call to {{{kernel()}}} or {{{left_kernel()}}} should arrive at the top of the hierarchy where it would take a transpose and call the (specialized) {{{right_kernel()}}}. So there wouldn't be a change in behavior in routines currently calling {{{kernel()}}} or {{{left_kernel()}}}, and Sage's preference for the left is retained by having the vanilla {{{kernel()}}} give back a left kernel. The speed-up for the computation of left kernels is up to 5% faster, and the computation of right kernels is up to 31% by eliminating paired transposes. The following timing statistics were obtained using sage.math: |
| Line 424: | Line 502: |
| * FIXME: summarize #5554 | * Cholesky decomposition for matrices other than {{{RDF}}} (Nick Alexander) -- The method {{{cholesky()}}} of the class {{{Matrix_double_dense}}} in {{{sage/matrix/matrix_double_dense.pyx}}} is now deprecated and will be removed in a future release. Users are advised to use {{{cholesky_decomposition()}}} instead. The new method {{{cholesky_decomposition()}}} in the class {{{Matrix}}} of {{{sage/matrix/matrix2.pyx}}} can be used to compute the Cholesky decomposition of matrices with entries over arbitrary precision real and complex fields. Here's an example over the real double field: {{{ sage: r = matrix(RDF, 5, 5, [ 0,0,0,0,1, 1,1,1,1,1, 16,8,4,2,1, 81,27,9,3,1, 256,64,16,4,1 ]) sage: m = r * r.transpose(); m [ 1.0 1.0 1.0 1.0 1.0] [ 1.0 5.0 31.0 121.0 341.0] [ 1.0 31.0 341.0 1555.0 4681.0] [ 1.0 121.0 1555.0 7381.0 22621.0] [ 1.0 341.0 4681.0 22621.0 69905.0] sage: L = m.cholesky_decomposition(); L [ 1.0 0.0 0.0 0.0 0.0] [ 1.0 2.0 0.0 0.0 0.0] [ 1.0 15.0 10.7238052948 0.0 0.0] [ 1.0 60.0 60.9858144589 7.79297342371 0.0] [ 1.0 170.0 198.623524155 39.3665667796 1.72309958068] sage: L.parent() Full MatrixSpace of 5 by 5 dense matrices over Real Double Field sage: L*L.transpose() [ 1.0 1.0 1.0 1.0 1.0] [ 1.0 5.0 31.0 121.0 341.0] [ 1.0 31.0 341.0 1555.0 4681.0] [ 1.0 121.0 1555.0 7381.0 22621.0] [ 1.0 341.0 4681.0 22621.0 69905.0] sage: ( L*L.transpose() - m ).norm(1) < 2^-30 True }}} Here's an example over a higher precision real field: {{{ sage: r = matrix(RealField(100), 5, 5, [ 0,0,0,0,1, 1,1,1,1,1, 16,8,4,2,1, 81,27,9,3,1, 256,64,16,4,1 ]) sage: m = r * r.transpose() sage: L = m.cholesky_decomposition() sage: L.parent() Full MatrixSpace of 5 by 5 dense matrices over Real Field with 100 bits of precision sage: ( L*L.transpose() - m ).norm(1) < 2^-50 True }}} Here's a Hermitian example: {{{ sage: r = matrix(CDF, 2, 2, [ 1, -2*I, 2*I, 6 ]); r [ 1.0 -2.0*I] [ 2.0*I 6.0] sage: r.eigenvalues() [0.298437881284, 6.70156211872] sage: ( r - r.conjugate().transpose() ).norm(1) < 1e-30 True sage: L = r.cholesky_decomposition(); L [ 1.0 0] [ 2.0*I 1.41421356237] sage: ( r - L*L.conjugate().transpose() ).norm(1) < 1e-30 True sage: L.parent() Full MatrixSpace of 2 by 2 dense matrices over Complex Double Field }}} Note that the implementation uses a standard recursion that is not known to be numerically stable. Furthermore, it is potentially expensive to ensure that the input is positive definite. Therefore this is not checked and it is possible that the output matrix is not a valid Cholesky decomposition of a matrix. * Make symbolic matrices use pynac symbolics (Mike Hansen, Jason Grout) -- Using Pynac symbolics, calculating the determinant of a symbolic matrix can be up to 2500x faster than previously. The following timing statistics were obtained using the machine sage.math: {{{ # BEFORE sage: x00, x01, x10, x11 = var("x00, x01, x10, x11") sage: a = matrix(2, [[x00,x01], [x10,x11]]) sage: %timeit a.det() 100 loops, best of 3: 8.29 ms per loop # AFTER sage: x00, x01, x10, x11 = var("x00, x01, x10, x11") sage: a = matrix(2, [[x00,x01], [x10,x11]]) sage: %timeit a.det() 100000 loops, best of 3: 3.2 µs per loop }}} |
| Line 435: | Line 590: |
* FIXME: summarize #5783 * FIXME: summarize #5796 * FIXME: summarize #5120 |
|
| Line 466: | Line 612: |
| * FIXME: summarize #6019 * FIXME: summarize #5924 == Notebook == |
* Slopes of {{{U_p}}} operator acting on a space of overconvergent modular forms (Lloyd Kilford) -- New method {{{slopes}}} of the class {{{OverconvergentModularFormsSpace}}} in {{{sage/modular/overconvergent/genus0.py}}} for computing the slopes of the {{{U_p}}} operator acting on a space of overconvergent modular forms. Here are some examples of using this new method: {{{ sage: OverconvergentModularForms(5,2,1/3,base_ring=Qp(5),prec=100).slopes(5) [0, 2, 5, 6, 9] sage: OverconvergentModularForms(2,1,1/3,char=DirichletGroup(4,QQ).0).slopes(5) [0, 2, 4, 6, 8] }}} |
| Line 476: | Line 623: |
| * FIXME: summarize #5250 * FIXME: summarize #6013 * FIXME: summarize #6008 * FIXME: summarize #6004 * FIXME: summarize #6059 * FIXME: summarize #6064 == Numerical == |
* Function {{{multiplicative_generator}}} for {{{Z/NZ}}} (David Loeffler) -- This adds support for the case where {{{n}}} is twice a power of an odd prime. Also, the new method {{{subgroups()}}} is added to the class {{{AbelianGroup_class}}} in {{{sage/groups/abelian_gps/abelian_group.py}}}. The method computes all the subgroups of a finite abelian group. Here's an example on working with the new method {{{subgroups()}}}: {{{ sage: AbelianGroup([2,3]).subgroups() [Multiplicative Abelian Group isomorphic to C2 x C3, which is the subgroup of Multiplicative Abelian Group isomorphic to C2 x C3 generated by [f0*f1^2], Multiplicative Abelian Group isomorphic to C2, which is the subgroup of Multiplicative Abelian Group isomorphic to C2 x C3 generated by [f0], Multiplicative Abelian Group isomorphic to C3, which is the subgroup of Multiplicative Abelian Group isomorphic to C2 x C3 generated by [f1], Trivial Abelian Group, which is the subgroup of Multiplicative Abelian Group isomorphic to C2 x C3 generated by []] sage: sage: len(AbelianGroup([2,3,8]).subgroups()) 22 sage: len(AbelianGroup([2,4,8]).subgroups()) 81 }}} * Speed-up relativization of number fields (Nick Alexander) -- The efficiency gain of relativizing a number field is up to 16%. Furthermore, the rewrite of the method {{{relativize()}}} allows for relativization over large number fields. The following timing statistics were obtained using the machine sage.math: {{{ # BEFORE sage: K.<a> = NumberField(x^10 - 2) sage: %time L.<c,d> = K.relativize(a^4 + a^2 + 2) CPU times: user 0.06 s, sys: 0.00 s, total: 0.06 s Wall time: 0.06 s #AFTER sage: K.<a> = NumberField(x^10 - 2) sage: %time L.<c,d> = K.relativize(a^4 + a^2 + 2) CPU times: user 0.05 s, sys: 0.01 s, total: 0.06 s Wall time: 0.05 s }}} * Improved efficiency of elliptic curve torsion computation (John Cremona) -- The speed-up of computing elliptic curve torsion can be up to 12%. The following timing statistics were obtained using the machine sage.math: {{{ # BEFORE sage: F.<z> = CyclotomicField(21) sage: E = EllipticCurve([2,-z^7,-z^7,0,0]) sage: time E._p_primary_torsion_basis(7); CPU times: user 9.87 s, sys: 0.07 s, total: 9.94 s Wall time: 9.95 s # AFTER sage: F.<z> = CyclotomicField(21) sage: E = EllipticCurve([2,-z^7,-z^7,0,0]) sage: time E._p_primary_torsion_basis(7,2); CPU times: user 8.56 s, sys: 0.11 s, total: 8.67 s Wall time: 8.67 s }}} * New method {{{odd_degree_model()}}} for hyperelliptic curves (Nick Alexander) -- The new method {{{odd_degree_model()}}} in the class {{{HyperellipticCurve_generic}}} of {{{sage/schemes/hyperelliptic_curves/hyperelliptic_generic.py}}} computes an odd degree model of a hyperelliptic curve. Here are some examples: {{{ sage: x = QQ['x'].gen() sage: H = HyperellipticCurve((x^2 + 2)*(x^2 + 3)*(x^2 + 5)) sage: K2 = QuadraticField(-2, 'a') sage: H.change_ring(K2).odd_degree_model() Hyperelliptic Curve over Number Field in a with defining polynomial x^2 + 2 defined by y^2 = 6*a*x^5 - 29*x^4 - 20*x^2 + 6*a*x + 1 sage: K3 = QuadraticField(-3, 'b') sage: H.change_ring(QuadraticField(-3, 'b')).odd_degree_model() Hyperelliptic Curve over Number Field in b with defining polynomial x^2 + 3 defined by y^2 = -4*b*x^5 - 14*x^4 - 20*b*x^3 - 35*x^2 + 6*b*x + 1 }}} * Rational arguments in {{{kronecker_symbol()}}} and {{{legendre_symbol()}}} (Gonzalo Tornaria) -- The functions {{{kronecker_symbol()}}} and {{{legendre_symbol()}}} in {{{sage/rings/arith.py}}} now support rational arguments. Here are some examples for working with rational arguments to these functions: {{{ sage: kronecker(2/3,5) 1 sage: legendre_symbol(2/3,7) -1 }}} |
| Line 494: | Line 712: |
| * FIXME: summarize #4223 * FIXME: summarize #6031 * FIXME: summarize #5934 * FIXME: summarize #1338 * FIXME: summarize #6032 * FIXME: summarize #6024 |
* Upgrade [[http://perso.ens-lyon.fr/damien.stehle/#software|fpLLL]] to latest upstream release version 3.0.12 (Michael Abshoff). * Update the [[http://www.shoup.net/ntl/|NTL]] spkg to version ntl-5.4.2.p7 (Michael Abshoff). * Downgrade the [[http://networkx.lanl.gov/|NetworkX]] spkg to version 0.36 (William Stein) -- The previous {{{networkx-0.99.p0.spkg}}} spkg contained both NetworkX 0.36, which Sage was using, and NetworkX 0.99. When installing {{{networkx-0.99.p0.spkg}}}, only version 0.36 would be installed. This wastes disk space, and it confuses users. The current NetworkX package that's shipped by Sage only contains version 0.36. * Upgrade [[http://www.algorithm.uni-bayreuth.de/en/research/SYMMETRICA/|Symmetrica]] to latest upstream release version 2.0 (Michael Abshoff). * Split off the [[http://www.boost.org/|Boost]] library from {{{polybori.spkg}}} (Michael Abshoff) -- Boost version 1.34.1 is now contained within its own spkg. * Switch from [[http://clisp.cons.org/|Clisp]] version 2.47 to [[http://ecls.sourceforge.net/|ECL]] version 9.4.1 (Michael Abshoff). * Upgrade [[http://www.mpir.org/|MPIR]] to latest upstream release version 1.1.2 (William Stein). * Upgrade [[http://www.python.org/|Python]] to latest 2.5.x upstream release version 2.5.4 (Michael Abshoff, Mike Hansen). |
| Line 509: | Line 739: |
| * FIXME: summarize #5105 * FIXME: summarize #5236 |
* Norm function in the {{{p}}}-adic ring (David Roe) -- New function {{{abs()}}} to calculate the {{{p}}}-adic absolute value. This is normalized so that the absolute value of {{{p}}} is {{{1/p}}}. This should be distinguished from the function {{{norm()}}}, which computes the norm of a {{{p}}}-adic element over a ground ring. Here are some examples of using the new function {{{abs()}}}: {{{ sage: a = Qp(5)(15); a.abs() 1/5 sage: a.abs(53) 0.200000000000000 sage: R = Zp(5, 5) sage: S.<x> = ZZ[] sage: f = x^5 + 75*x^3 - 15*x^2 +125*x - 5 sage: W.<w> = R.ext(f) sage: w.abs() 0.724779663677696 }}} |
| Line 517: | Line 758: |
| * FIXME: summarize #6037 | * Major upgrade to the {{{QuadraticForm}}} local density routines (Jon Hanke) -- A complete rewrite of local densities routines, following a consistent interface (and algorithms) as described in [[http://trac.sagemath.org/sage_trac/attachment/ticket/6037/Local%20Densities%20Writeup.pdf|this paper]]. |
| Line 523: | Line 764: |
| * FIXME: summarize #5777 * FIXME: summarize #5930 |
* Update [[http://pynac.sagemath.org/|Pynac]] to version 0.1.7 (Burcin Erocal). * Switch from Maxima to Pynac for core symbolic manipulation (Mike Hansen, William Stein, Carl Witty, Robert Bradshaw). |
| Line 536: | Line 778: |
== User Interface == == Website / Wiki == |
Sage 4.0 Release Tour
Sage 4.0 was released on May 29, 2009. For the official, comprehensive release note, please refer to sage-4.0.txt. A nicely formatted version of this release tour can be found at this Wordpress blog. The following points are some of the foci of this release:
- New symbolics based on Pynac
- Bring doctest coverage up to at least 75%
- Solaris 10 support (at least for gcc 4.3.x + gmake)
- Switch from Clisp to ECL
- gcc 4.4.0 support
- OS X 64-bit support
Algebra
Deprecate the order() method on elements of rings (John Palmieri) -- The method order() of the class sage.structure.element.RingElement is now deprecated and will be removed in a future release. For additive or multiplicative order, use the additive_order or multiplicative_order method respectively.
Partial fraction decomposition for irreducible denominators (Gonzalo Tornaria) -- For example, over the field ZZ[x] you can do
sage: R.<x> = ZZ["x"] sage: q = x^2 / (x - 1) sage: q.partial_fraction_decomposition() (x + 1, [1/(x - 1)]) sage: q = x^10 / (x - 1)^5 sage: whole, parts = q.partial_fraction_decomposition() sage: whole + sum(parts) x^10/(x^5 - 5*x^4 + 10*x^3 - 10*x^2 + 5*x - 1) sage: whole + sum(parts) == q True
and over the finite field GF(2)[x]:
sage: R.<x> = GF(2)["x"] sage: q = (x + 1) / (x^3 + x + 1) qsage: q.partial_fraction_decomposition() (0, [(x + 1)/(x^3 + x + 1)])
Algebraic Geometry
Various invariants for genus 2 hyperelliptic curves (Nick Alexander) -- The following invariants for genus 2 hyperelliptic curves are implemented in the module sage/schemes/hyperelliptic_curves/hyperelliptic_g2_generic.py:
- the Clebsch invariants
- the Igusa-Clebsch invariants
- the absolute Igusa invariants
Basic Arithmetic
Utility methods for integer arithmetics (Fredrik Johansson) -- New methods trailing_zero_bits() and sqrtrem() for the class Integer in sage/rings/integer.pyx:
trailing_zero_bits(self) -- Returns the number of trailing zero bits in self, i.e. the exponent of the largest power of 2 dividing self.
sqrtrem(self) -- Returns a pair (s, r) where s is the integer square root of self and r is the remainder such that self = s^2 + r.
sage: 13.trailing_zero_bits() 0 sage: (-13).trailing_zero_bits() 0 sage: (-13 >> 2).trailing_zero_bits() 2 sage: (-13 >> 3).trailing_zero_bits() 1 sage: (-13 << 3).trailing_zero_bits() 3 sage: (-13 << 2).trailing_zero_bits() 2 sage: 29.sqrtrem() (5, 4) sage: 25.sqrtrem() (5, 0)
Coercion
Coercion from float to rationals (Robert Bradshaw) -- One can now coerce a number of type float to QQ. Here's an example:
sage: a = float(1.0) sage: QQ(a) 1 sage: type(a); type(QQ(a)) <type 'float'> <type 'sage.rings.rational.Rational'>
Combinatorics
ASCII art output for Dynkin diagrams (Dan Bump) -- Support for ASCII art representation of Dynkin diagrams of a finite Cartan type. Here are some examples:
sage: DynkinDiagram("E6") O 2 | | O---O---O---O---O 1 3 4 5 6 E6 sage: DynkinDiagram(['E',6,1]) O 0 | | O 2 | | O---O---O---O---O 1 3 4 5 6 E6~Crystal of letters for type E (Brant Jones, Anne Schilling) -- Support crystal of letters for type E corresponding to the highest weight crystal B(\Lambda_1) and its dual B(\Lambda_6) (using the Sage labeling convention of the Dynkin nodes). Here are some examples:
sage: C = CrystalOfLetters(['E',6]) sage: C.list() [[1], [-1, 3], [-3, 4], [-4, 2, 5], [-2, 5], [-5, 2, 6], [-2, -5, 4, 6], [-4, 3, 6], [-3, 1, 6], [-1, 6], [-6, 2], [-2, -6, 4], [-4, -6, 3, 5], [-3, -6, 1, 5], [-1, -6, 5], [-5, 3], [-3, -5, 1, 4], [-1, -5, 4], [-4, 1, 2], [-1, -4, 2, 3], [-3, 2], [-2, -3, 4], [-4, 5], [-5, 6], [-6], [-2, 1], [-1, -2, 3]] sage: C = CrystalOfLetters(['E',6], element_print_style="compact") sage: C.list() [+, a, b, c, d, e, f, g, h, i, j, k, l, m, n, o, p, q, r, s, t, u, v, w, x, y, z]
Commutative Algebra
Improved performance for SR (Martin Albrecht) -- The speed-up gain for SR is up to 6x. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: sr = mq.SR(4, 4, 4, 8, gf2=True, polybori=True, allow_zero_inversions=True) sage: %time F,s = sr.polynomial_system() CPU times: user 21.65 s, sys: 0.03 s, total: 21.68 s Wall time: 21.83 s # AFTER sage: sr = mq.SR(4, 4, 4, 8, gf2=True, polybori=True, allow_zero_inversions=True) sage: %time F,s = sr.polynomial_system() CPU times: user 3.61 s, sys: 0.06 s, total: 3.67 s Wall time: 3.67 s
Symmetric Groebner bases and infinitely generated polynomial rings (Simon King, Mike Hansen) -- The new modules sage/rings/polynomial/infinite_polynomial_element.py and sage/rings/polynomial/infinite_polynomial_ring.py support computation in polynomial rings with a countably infinite number of variables. Here are some examples for working with these new modules:
sage: from sage.rings.polynomial.infinite_polynomial_element import InfinitePolynomial sage: X.<x> = InfinitePolynomialRing(QQ) sage: a = InfinitePolynomial(X, "(x1 + x2)^2"); a x2^2 + 2*x2*x1 + x1^2 sage: p = a.polynomial() sage: b = InfinitePolynomial(X, a.polynomial()) sage: a == b True sage: InfinitePolynomial(X, int(1)) 1 sage: InfinitePolynomial(X, 1) 1 sage: Y.<x,y> = InfinitePolynomialRing(GF(2), implementation="sparse") sage: InfinitePolynomial(Y, a) x2^2 + x1^2 sage: X.<x,y> = InfinitePolynomialRing(QQ, implementation="sparse") sage: A.<a,b> = InfinitePolynomialRing(QQ, order="deglex") sage: f = x[5] + 2; f x5 + 2 sage: g = 3*y[1]; g 3*y1 sage: g._p.parent() Univariate Polynomial Ring in y1 over Rational Field sage: f2 = a[5] + 2; f2 a5 + 2 sage: g2 = 3*b[1]; g2 3*b1 sage: A.polynomial_ring() Multivariate Polynomial Ring in b5, b4, b3, b2, b1, b0, a5, a4, a3, a2, a1, a0 over Rational Field sage: f + g 3*y1 + x5 + 2 sage: p = x[10]^2 * (f + g); p 3*y1*x10^2 + x10^2*x5 + 2*x10^2
Furthermore, the new module sage/rings/polynomial/symmetric_ideal.py supports ideals of polynomial rings in a countably infinite number of variables that are invariant under variable permutation. Symmetric reduction of infinite polynomials is provided by the new module sage/rings/polynomial/symmetric_reduction.pyx.
Geometry
Simplicial complex method for polytopes (Marshall Hampton) -- New method simplicial_complex() in the class Polyhedron of sage/geometry/polyhedra.py for computing the simplicial complex from a triangulation of the polytope. Here's an example:
sage: p = polytopes.cuboctahedron() sage: p.simplicial_complex() Simplicial complex with 13 vertices and 20 facets
Face lattices and f-vectors for polytopes (Marshall Hampton) -- New methods face_lattice() and f_vector() in the class Polyhedron of sage/geometry/polyhedra.py:
face_lattice() -- Returns the face-lattice poset. Elements are tuples of (vertices, facets) which keeps track of both the vertices in each face, and all the facets containing them. This method implements the results from the following paper:
- V. Kaibel and M.E. Pfetsch. Computing the face lattice of a polytope from its vertex-facet incidences. Computational Geometry, 23(3):281--290, 2002.
f_vector() -- Returns the f-vector of a polytope as a list.
sage: c5_10 = Polyhedron(vertices = [[i,i^2,i^3,i^4,i^5] for i in xrange(1,11)]) sage: c5_10_fl = c5_10.face_lattice() sage: [len(x) for x in c5_10_fl.level_sets()] [1, 10, 45, 100, 105, 42, 1] sage: p = Polyhedron(vertices = [[1, 2, 3], [1, 3, 2], [2, 1, 3], [2, 3, 1], [3, 1, 2], [3, 2, 1], [0, 0, 0]]) sage: p.f_vector() [1, 7, 12, 7, 1]
Graph Theory
Graph colouring (Robert Miller) -- New method coloring() of the class sage.graphs.graph.Graph for obtaining the first (optimal) coloring found on a graph. Here are some examples on using this new method:
sage: G = Graph("Fooba") sage: P = G.coloring() sage: G.plot(partition=P) sage: H = G.coloring(hex_colors=True) sage: G.plot(vertex_colors=H)
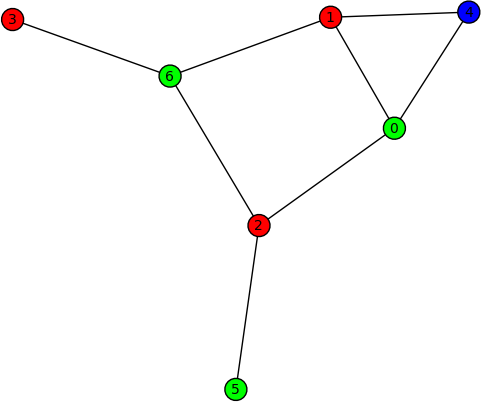
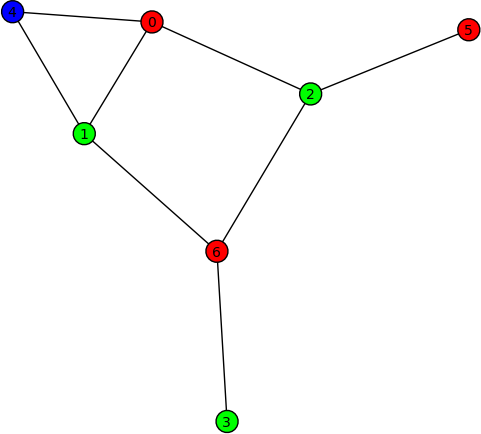
- Optimize the construction of large Sage graphs (Radoslav Kirov) -- The construction of large Sage graphs is now up to 19x faster than previously. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: D = {} sage: for i in xrange(10^3): ....: D[i] = [i+1, i-1] ....: sage: timeit("g = Graph(D)") 5 loops, best of 3: 1.02 s per loop # AFTER sage: D = {} sage: for i in xrange(10^3): ....: D[i] = [i+1, i-1] ....: sage: timeit("g = Graph(D)") 5 loops, best of 3: 51.2 ms per loop Generate size n trees in linear time (Ryan Dingman) -- The speed-up can be up to 3400x. However, the efficiency gain is greater as n becomes larger. The following timing statistics were produced using the machine sage.math:
# BEFORE sage: %time L = list(graphs.trees(2)) CPU times: user 0.13 s, sys: 0.02 s, total: 0.15 s Wall time: 0.18 s sage: %time L = list(graphs.trees(4)) CPU times: user 0.02 s, sys: 0.00 s, total: 0.02 s Wall time: 0.02 s sage: %time L = list(graphs.trees(6)) CPU times: user 0.08 s, sys: 0.00 s, total: 0.08 s Wall time: 0.07 s sage: %time L = list(graphs.trees(8)) CPU times: user 0.59 s, sys: 0.00 s, total: 0.59 s Wall time: 0.60 s sage: %time L = list(graphs.trees(10)) CPU times: user 34.48 s, sys: 0.02 s, total: 34.50 s Wall time: 34.51 s # AFTER sage: %time L = list(graphs.trees(2)) CPU times: user 0.11 s, sys: 0.02 s, total: 0.13 s Wall time: 0.15 s sage: %time L = list(graphs.trees(4)) CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.00 s sage: %time L = list(graphs.trees(6)) CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s Wall time: 0.00 s sage: %time L = list(graphs.trees(8)) CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s Wall time: 0.00 s sage: %time L = list(graphs.trees(10)) CPU times: user 0.01 s, sys: 0.00 s, total: 0.01 s Wall time: 0.01 s sage: %time L = list(graphs.trees(12)) CPU times: user 0.06 s, sys: 0.00 s, total: 0.06 s Wall time: 0.05 s sage: %time L = list(graphs.trees(14)) CPU times: user 0.51 s, sys: 0.01 s, total: 0.52 s Wall time: 0.52 s
Graphics
Implicit Surfaces (Bill Cauchois, Carl Witty) -- New function implicit_plot3d for plotting level sets of 3-D functions. The documentation contains many examples. Here's a sphere contained inside a tube-like sphere:
sage: x, y, z = var("x, y, z") sage: T = 1.61803398875 sage: p = 2 - (cos(x + T*y) + cos(x - T*y) + cos(y + T*z) + cos(y - T*z) + cos(z - T*x) + cos(z + T*x)) sage: r = 4.77 sage: implicit_plot3d(p, (-r, r), (-r, r), (-r, r), plot_points=40, zoom=1.2).show()

- Here's a Klein bottle:
sage: x, y, z = var("x, y, z") sage: implicit_plot3d((x^2+y^2+z^2+2*y-1)*((x^2+y^2+z^2-2*y-1)^2-8*z^2)+16*x*z*(x^2+y^2+z^2-2*y-1), (x, -3, 3), (y, -3.1, 3.1), (z, -4, 4), zoom=1.2)
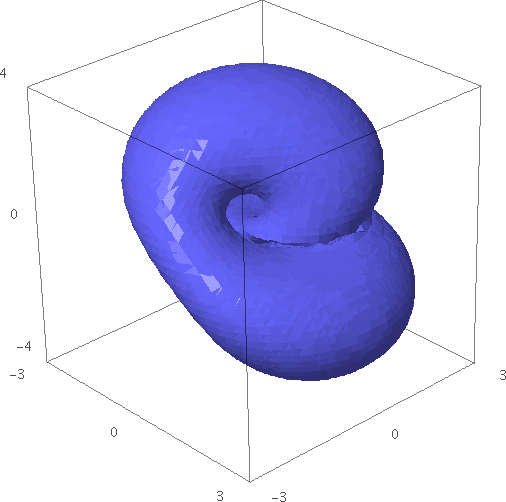
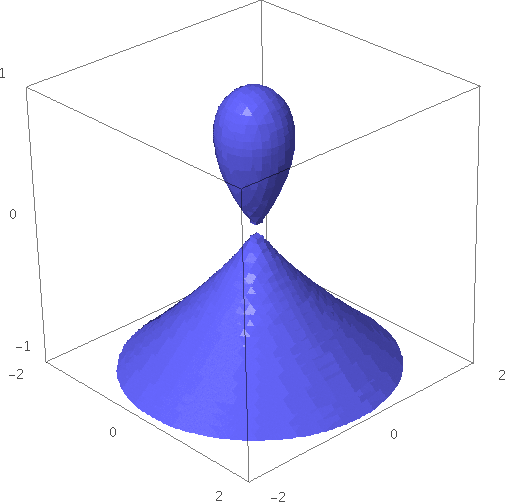
- This example shows something resembling a water droplet:
sage: x, y, z = var("x, y, z") sage: implicit_plot3d(x^2 +y^2 -(1-z)*z^2, (x, -1.5, 1.5), (y, -1.5, 1.5), (z, -1, 1), zoom=1.2)
- Fixed bug in rendering 2D polytopes embedded in 3D (Arnauld Bergeron, Bill Cauchois, Marshall Hampton).
Group Theory
Improved efficiency of is_subgroup (Simon King) -- Testing whether a group is a subgroup of another group is now up to 2x faster than previously. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: G = SymmetricGroup(7) sage: H = SymmetricGroup(6) sage: %time H.is_subgroup(G) CPU times: user 4.12 s, sys: 0.53 s, total: 4.65 s Wall time: 5.51 s True sage: %timeit H.is_subgroup(G) 10000 loops, best of 3: 118 µs per loop # AFTER sage: G = SymmetricGroup(7) sage: H = SymmetricGroup(6) sage: %time H.is_subgroup(G) CPU times: user 0.00 s, sys: 0.00 s, total: 0.00 s Wall time: 0.00 s True sage: %timeit H.is_subgroup(G) 10000 loops, best of 3: 56.3 µs per loop
Interfaces
Viewing Sage objects with a PDF viewer (Nicolas Thiery) -- Implements the option viewer="pdf" for the command view() so that one can invoke this command in the form view(object, viewer="pdf") in order to view object using a PDF viewer. Typical uses of this new optional argument include:
- You prefer to use a PDF viewer rather than a DVI viewer.
- You want to view LaTeX snippets which are not displayed well in DVI viewers (e.g. graphics produced using tikzpicture).
Change name of Pari's sum function when imported (Craig Citro) -- When Pari's sum function is imported, it is renamed to pari_sum in order to avoid conflict Python's sum function.
Linear Algebra
Improved performance for the generic linear_combination_of_rows and linear_combination_of_columns functions for matrices (William Stein) -- The speed-up for the generic functions linear_combination_of_rows and linear_combination_of_columns is up to 4x. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: A = random_matrix(QQ, 50) sage: v = [1..50] sage: %timeit A.linear_combination_of_rows(v); 1000 loops, best of 3: 1.99 ms per loop sage: %timeit A.linear_combination_of_columns(v); 1000 loops, best of 3: 1.97 ms per loop # AFTER sage: A = random_matrix(QQ, 50) sage: v = [1..50] sage: %timeit A.linear_combination_of_rows(v); 1000 loops, best of 3: 436 µs per loop sage: %timeit A.linear_combination_of_columns(v); 1000 loops, best of 3: 457 µs per loop
Massively improved performance for 4 x 4 determinants (Tom Boothby) -- The efficiency of computing the determinants of 4 x 4 matrices can range from 16x up to 58,083x faster than previously, depending on the base ring. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: S = MatrixSpace(ZZ, 4) sage: M = S.random_element(1, 10^8) sage: timeit("M.det(); M._clear_cache()") 625 loops, best of 3: 53 µs per loop sage: M = S.random_element(1, 10^10) sage: timeit("M.det(); M._clear_cache()") 625 loops, best of 3: 54.1 µs per loop sage: sage: M = S.random_element(1, 10^200) sage: timeit("M.det(); M._clear_cache()") 5 loops, best of 3: 121 ms per loop sage: M = S.random_element(1, 10^300) sage: timeit("M.det(); M._clear_cache()") 5 loops, best of 3: 338 ms per loop sage: M = S.random_element(1, 10^1000) sage: timeit("M.det(); M._clear_cache()") 5 loops, best of 3: 9.7 s per loop # AFTER sage: S = MatrixSpace(ZZ, 4) sage: M = S.random_element(1, 10^8) sage: timeit("M.det(); M._clear_cache()") 625 loops, best of 3: 3.17 µs per loop sage: M = S.random_element(1, 10^10) sage: timeit("M.det(); M._clear_cache()") 625 loops, best of 3: 3.44 µs per loop sage: sage: M = S.random_element(1, 10^200) sage: timeit("M.det(); M._clear_cache()") 625 loops, best of 3: 15.3 µs per loop sage: M = S.random_element(1, 10^300) sage: timeit("M.det(); M._clear_cache()") 625 loops, best of 3: 27 µs per loop sage: M = S.random_element(1, 10^1000) sage: timeit("M.det(); M._clear_cache()") 625 loops, best of 3: 167 µs per loopRefactor matrix kernels (Rob Beezer) -- The core section of kernel computation for each (specialized) class is now moved into the method right_kernel(). Mostly these would replace kernel() methods that are computing left kernels. A call to kernel() or left_kernel() should arrive at the top of the hierarchy where it would take a transpose and call the (specialized) right_kernel(). So there wouldn't be a change in behavior in routines currently calling kernel() or left_kernel(), and Sage's preference for the left is retained by having the vanilla kernel() give back a left kernel. The speed-up for the computation of left kernels is up to 5% faster, and the computation of right kernels is up to 31% by eliminating paired transposes. The following timing statistics were obtained using sage.math:
# BEFORE sage: n = 2000 sage: entries = [[1/(i+j+1) for i in srange(n)] for j in srange(n)] sage: mat = matrix(QQ, entries) sage: %time mat.left_kernel(); CPU times: user 21.92 s, sys: 3.22 s, total: 25.14 s Wall time: 25.26 s sage: %time mat.right_kernel(); CPU times: user 23.62 s, sys: 3.32 s, total: 26.94 s Wall time: 26.94 s # AFTER sage: n = 2000 sage: entries = [[1/(i+j+1) for i in srange(n)] for j in srange(n)] sage: mat = matrix(QQ, entries) sage: %time mat.left_kernel(); CPU times: user 20.87 s, sys: 2.94 s, total: 23.81 s Wall time: 23.89 s sage: %time mat.right_kernel(); CPU times: user 18.43 s, sys: 0.00 s, total: 18.43 s Wall time: 18.43 s
Cholesky decomposition for matrices other than RDF (Nick Alexander) -- The method cholesky() of the class Matrix_double_dense in sage/matrix/matrix_double_dense.pyx is now deprecated and will be removed in a future release. Users are advised to use cholesky_decomposition() instead. The new method cholesky_decomposition() in the class Matrix of sage/matrix/matrix2.pyx can be used to compute the Cholesky decomposition of matrices with entries over arbitrary precision real and complex fields. Here's an example over the real double field:
sage: r = matrix(RDF, 5, 5, [ 0,0,0,0,1, 1,1,1,1,1, 16,8,4,2,1, 81,27,9,3,1, 256,64,16,4,1 ]) sage: m = r * r.transpose(); m [ 1.0 1.0 1.0 1.0 1.0] [ 1.0 5.0 31.0 121.0 341.0] [ 1.0 31.0 341.0 1555.0 4681.0] [ 1.0 121.0 1555.0 7381.0 22621.0] [ 1.0 341.0 4681.0 22621.0 69905.0] sage: L = m.cholesky_decomposition(); L [ 1.0 0.0 0.0 0.0 0.0] [ 1.0 2.0 0.0 0.0 0.0] [ 1.0 15.0 10.7238052948 0.0 0.0] [ 1.0 60.0 60.9858144589 7.79297342371 0.0] [ 1.0 170.0 198.623524155 39.3665667796 1.72309958068] sage: L.parent() Full MatrixSpace of 5 by 5 dense matrices over Real Double Field sage: L*L.transpose() [ 1.0 1.0 1.0 1.0 1.0] [ 1.0 5.0 31.0 121.0 341.0] [ 1.0 31.0 341.0 1555.0 4681.0] [ 1.0 121.0 1555.0 7381.0 22621.0] [ 1.0 341.0 4681.0 22621.0 69905.0] sage: ( L*L.transpose() - m ).norm(1) < 2^-30 True
Here's an example over a higher precision real field:sage: r = matrix(RealField(100), 5, 5, [ 0,0,0,0,1, 1,1,1,1,1, 16,8,4,2,1, 81,27,9,3,1, 256,64,16,4,1 ]) sage: m = r * r.transpose() sage: L = m.cholesky_decomposition() sage: L.parent() Full MatrixSpace of 5 by 5 dense matrices over Real Field with 100 bits of precision sage: ( L*L.transpose() - m ).norm(1) < 2^-50 True
Here's a Hermitian example:sage: r = matrix(CDF, 2, 2, [ 1, -2*I, 2*I, 6 ]); r [ 1.0 -2.0*I] [ 2.0*I 6.0] sage: r.eigenvalues() [0.298437881284, 6.70156211872] sage: ( r - r.conjugate().transpose() ).norm(1) < 1e-30 True sage: L = r.cholesky_decomposition(); L [ 1.0 0] [ 2.0*I 1.41421356237] sage: ( r - L*L.conjugate().transpose() ).norm(1) < 1e-30 True sage: L.parent() Full MatrixSpace of 2 by 2 dense matrices over Complex Double Field
Note that the implementation uses a standard recursion that is not known to be numerically stable. Furthermore, it is potentially expensive to ensure that the input is positive definite. Therefore this is not checked and it is possible that the output matrix is not a valid Cholesky decomposition of a matrix.- Make symbolic matrices use pynac symbolics (Mike Hansen, Jason Grout) -- Using Pynac symbolics, calculating the determinant of a symbolic matrix can be up to 2500x faster than previously. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: x00, x01, x10, x11 = var("x00, x01, x10, x11") sage: a = matrix(2, [[x00,x01], [x10,x11]]) sage: %timeit a.det() 100 loops, best of 3: 8.29 ms per loop # AFTER sage: x00, x01, x10, x11 = var("x00, x01, x10, x11") sage: a = matrix(2, [[x00,x01], [x10,x11]]) sage: %timeit a.det() 100000 loops, best of 3: 3.2 µs per loop
Miscellaneous
Allow use of pdflatex instead of latex (John Palmieri) -- One can now use pdflatex instead of latex in two different ways:
Use a %pdflatex cell in a notebook; or
Call latex.pdflatex(True)
after which any use of latex (in a %latex cell or using the view command) will use pdflatex. One visually appealing aspect of this is that if you have the most recent version of pgf installed, as well as the tkz-graph package, you can produce images like the following:
Modular Forms
Action of Hecke operators on Gamma_1(N) modular forms (David Loeffler) -- Here's an example:
sage: ModularForms(Gamma1(11), 2).hecke_matrix(2) [ -2 0 0 0 0 0 0 0 0 0] [ 0 -381 0 -360 0 120 -4680 -6528 -1584 7752] [ 0 -190 0 -180 0 60 -2333 -3262 -789 3887] [ 0 -634/11 1 -576/11 0 170/11 -7642/11 -10766/11 -231 12555/11] [ 0 98/11 0 78/11 0 -26/11 1157/11 1707/11 30 -1959/11] [ 0 290/11 0 271/11 0 -50/11 3490/11 5019/11 99 -5694/11] [ 0 230/11 0 210/11 0 -70/11 2807/11 3940/11 84 -4632/11] [ 0 122/11 0 120/11 1 -40/11 1505/11 2088/11 48 -2463/11] [ 0 42/11 0 46/11 0 -30/11 554/11 708/11 21 -970/11] [ 0 10/11 0 12/11 0 7/11 123/11 145/11 7 -177/11]
Slopes of U_p operator acting on a space of overconvergent modular forms (Lloyd Kilford) -- New method slopes of the class OverconvergentModularFormsSpace in sage/modular/overconvergent/genus0.py for computing the slopes of the U_p operator acting on a space of overconvergent modular forms. Here are some examples of using this new method:
sage: OverconvergentModularForms(5,2,1/3,base_ring=Qp(5),prec=100).slopes(5) [0, 2, 5, 6, 9] sage: OverconvergentModularForms(2,1,1/3,char=DirichletGroup(4,QQ).0).slopes(5) [0, 2, 4, 6, 8]
Number Theory
Function multiplicative_generator for Z/NZ (David Loeffler) -- This adds support for the case where n is twice a power of an odd prime. Also, the new method subgroups() is added to the class AbelianGroup_class in sage/groups/abelian_gps/abelian_group.py. The method computes all the subgroups of a finite abelian group. Here's an example on working with the new method subgroups():
sage: AbelianGroup([2,3]).subgroups() [Multiplicative Abelian Group isomorphic to C2 x C3, which is the subgroup of Multiplicative Abelian Group isomorphic to C2 x C3 generated by [f0*f1^2], Multiplicative Abelian Group isomorphic to C2, which is the subgroup of Multiplicative Abelian Group isomorphic to C2 x C3 generated by [f0], Multiplicative Abelian Group isomorphic to C3, which is the subgroup of Multiplicative Abelian Group isomorphic to C2 x C3 generated by [f1], Trivial Abelian Group, which is the subgroup of Multiplicative Abelian Group isomorphic to C2 x C3 generated by []] sage: sage: len(AbelianGroup([2,3,8]).subgroups()) 22 sage: len(AbelianGroup([2,4,8]).subgroups()) 81
Speed-up relativization of number fields (Nick Alexander) -- The efficiency gain of relativizing a number field is up to 16%. Furthermore, the rewrite of the method relativize() allows for relativization over large number fields. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: K.<a> = NumberField(x^10 - 2) sage: %time L.<c,d> = K.relativize(a^4 + a^2 + 2) CPU times: user 0.06 s, sys: 0.00 s, total: 0.06 s Wall time: 0.06 s #AFTER sage: K.<a> = NumberField(x^10 - 2) sage: %time L.<c,d> = K.relativize(a^4 + a^2 + 2) CPU times: user 0.05 s, sys: 0.01 s, total: 0.06 s Wall time: 0.05 s
- Improved efficiency of elliptic curve torsion computation (John Cremona) -- The speed-up of computing elliptic curve torsion can be up to 12%. The following timing statistics were obtained using the machine sage.math:
# BEFORE sage: F.<z> = CyclotomicField(21) sage: E = EllipticCurve([2,-z^7,-z^7,0,0]) sage: time E._p_primary_torsion_basis(7); CPU times: user 9.87 s, sys: 0.07 s, total: 9.94 s Wall time: 9.95 s # AFTER sage: F.<z> = CyclotomicField(21) sage: E = EllipticCurve([2,-z^7,-z^7,0,0]) sage: time E._p_primary_torsion_basis(7,2); CPU times: user 8.56 s, sys: 0.11 s, total: 8.67 s Wall time: 8.67 s
New method odd_degree_model() for hyperelliptic curves (Nick Alexander) -- The new method odd_degree_model() in the class HyperellipticCurve_generic of sage/schemes/hyperelliptic_curves/hyperelliptic_generic.py computes an odd degree model of a hyperelliptic curve. Here are some examples:
sage: x = QQ['x'].gen() sage: H = HyperellipticCurve((x^2 + 2)*(x^2 + 3)*(x^2 + 5)) sage: K2 = QuadraticField(-2, 'a') sage: H.change_ring(K2).odd_degree_model() Hyperelliptic Curve over Number Field in a with defining polynomial x^2 + 2 defined by y^2 = 6*a*x^5 - 29*x^4 - 20*x^2 + 6*a*x + 1 sage: K3 = QuadraticField(-3, 'b') sage: H.change_ring(QuadraticField(-3, 'b')).odd_degree_model() Hyperelliptic Curve over Number Field in b with defining polynomial x^2 + 3 defined by y^2 = -4*b*x^5 - 14*x^4 - 20*b*x^3 - 35*x^2 + 6*b*x + 1
Rational arguments in kronecker_symbol() and legendre_symbol() (Gonzalo Tornaria) -- The functions kronecker_symbol() and legendre_symbol() in sage/rings/arith.py now support rational arguments. Here are some examples for working with rational arguments to these functions:
sage: kronecker(2/3,5) 1 sage: legendre_symbol(2/3,7) -1
Packages
Upgrade fpLLL to latest upstream release version 3.0.12 (Michael Abshoff).
Update the NTL spkg to version ntl-5.4.2.p7 (Michael Abshoff).
Downgrade the NetworkX spkg to version 0.36 (William Stein) -- The previous networkx-0.99.p0.spkg spkg contained both NetworkX 0.36, which Sage was using, and NetworkX 0.99. When installing networkx-0.99.p0.spkg, only version 0.36 would be installed. This wastes disk space, and it confuses users. The current NetworkX package that's shipped by Sage only contains version 0.36.
Upgrade Symmetrica to latest upstream release version 2.0 (Michael Abshoff).
Split off the Boost library from polybori.spkg (Michael Abshoff) -- Boost version 1.34.1 is now contained within its own spkg.
Switch from Clisp version 2.47 to ECL version 9.4.1 (Michael Abshoff).
Upgrade MPIR to latest upstream release version 1.1.2 (William Stein).
Upgrade Python to latest 2.5.x upstream release version 2.5.4 (Michael Abshoff, Mike Hansen).
P-adics
Norm function in the p-adic ring (David Roe) -- New function abs() to calculate the p-adic absolute value. This is normalized so that the absolute value of p is 1/p. This should be distinguished from the function norm(), which computes the norm of a p-adic element over a ground ring. Here are some examples of using the new function abs():
sage: a = Qp(5)(15); a.abs() 1/5 sage: a.abs(53) 0.200000000000000 sage: R = Zp(5, 5) sage: S.<x> = ZZ[] sage: f = x^5 + 75*x^3 - 15*x^2 +125*x - 5 sage: W.<w> = R.ext(f) sage: w.abs() 0.724779663677696
Quadratic Forms
Major upgrade to the QuadraticForm local density routines (Jon Hanke) -- A complete rewrite of local densities routines, following a consistent interface (and algorithms) as described in this paper.
Symbolics
Update Pynac to version 0.1.7 (Burcin Erocal).
- Switch from Maxima to Pynac for core symbolic manipulation (Mike Hansen, William Stein, Carl Witty, Robert Bradshaw).
Topology
Random simplicial complexes (John Palmieri) -- New method RandomComplex() in the module sage/homology/examples.py for producing a random d-dimensional simplicial complex on n vertices. Here's an example:
sage: simplicial_complexes.RandomComplex(6,12) Simplicial complex with vertex set (0, 1, 2, 3, 4, 5, 6, 7) and facets {(0, 1, 2, 3, 4, 5, 6, 7)}
